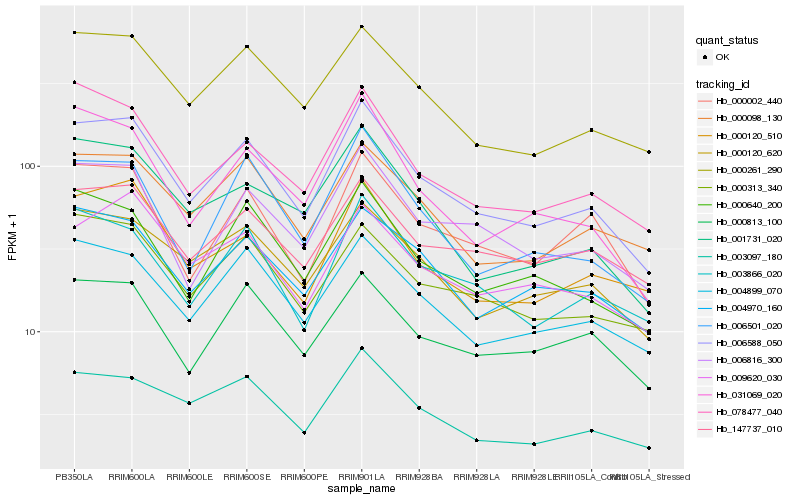
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_050 |
0.0 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 2 |
Hb_000261_290 |
0.069698069 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 3 |
Hb_004899_070 |
0.0748211035 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 4 |
Hb_003866_020 |
0.0779931071 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006816_300 |
0.0781153257 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 6 |
Hb_000120_620 |
0.078643606 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 7 |
Hb_003097_180 |
0.0788478157 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 8 |
Hb_000640_200 |
0.0806372786 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 9 |
Hb_000098_130 |
0.0807541366 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 10 |
Hb_004970_160 |
0.0812350261 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000313_340 |
0.0818090018 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 12 |
Hb_006501_020 |
0.0823406626 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 13 |
Hb_000002_440 |
0.0830127759 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 14 |
Hb_078477_040 |
0.0850029882 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 15 |
Hb_031069_020 |
0.0854561829 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |
| 16 |
Hb_001731_020 |
0.0861813705 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 17 |
Hb_147737_010 |
0.0863061044 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000120_510 |
0.0868343933 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000813_100 |
0.0868780921 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 20 |
Hb_009620_030 |
0.0886562397 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |