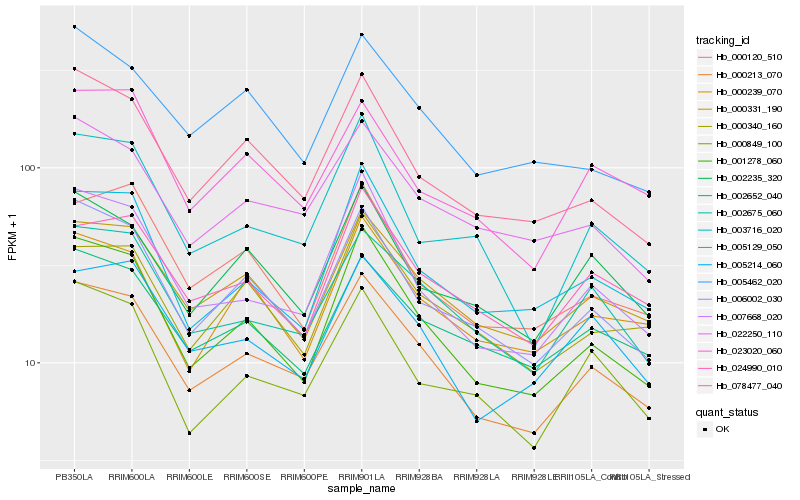
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000213_070 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001278_060 |
0.0619151706 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 3 |
Hb_006002_030 |
0.0689724753 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000331_190 |
0.0712358779 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 5 |
Hb_002675_060 |
0.0727063722 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007668_020 |
0.0746362264 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 7 |
Hb_078477_040 |
0.0774530409 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 8 |
Hb_022250_110 |
0.079908431 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_005214_060 |
0.0826955451 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_002235_320 |
0.0859460289 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002652_040 |
0.0877958157 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 12 |
Hb_000849_100 |
0.0888359617 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 13 |
Hb_023020_060 |
0.0890299222 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 14 |
Hb_000340_160 |
0.0903659608 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 15 |
Hb_005129_050 |
0.0904972154 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase [Jatropha curcas] |
| 16 |
Hb_024990_010 |
0.0913696049 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 17 |
Hb_000120_510 |
0.0918038197 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003716_020 |
0.091887335 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 19 |
Hb_005462_020 |
0.0919207615 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 20 |
Hb_000239_070 |
0.0921134563 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |