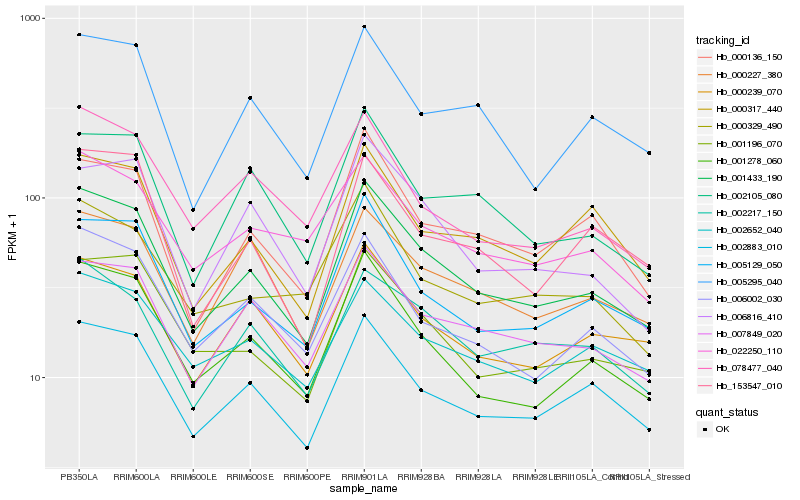
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_190 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 2 |
Hb_001278_060 |
0.0662971676 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 3 |
Hb_005129_050 |
0.0680089687 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase [Jatropha curcas] |
| 4 |
Hb_002883_010 |
0.068063761 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007849_020 |
0.0805135018 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 6 |
Hb_001196_070 |
0.0808204248 |
- |
- |
nucleolar protein nop56, putative [Ricinus communis] |
| 7 |
Hb_006002_030 |
0.082223969 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000136_150 |
0.0846064078 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 9 |
Hb_002217_150 |
0.0864012023 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 10 |
Hb_000239_070 |
0.0865295176 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000317_440 |
0.0867696466 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005295_040 |
0.0878624306 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 13 |
Hb_002105_080 |
0.0894983194 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 14 |
Hb_006816_410 |
0.0913153549 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 15 |
Hb_022250_110 |
0.0943282493 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_078477_040 |
0.0947348824 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 17 |
Hb_153547_010 |
0.0948022328 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 18 |
Hb_002652_040 |
0.0952365115 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 19 |
Hb_000227_380 |
0.0957499534 |
- |
- |
PREDICTED: protein TIME FOR COFFEE isoform X3 [Jatropha curcas] |
| 20 |
Hb_000329_490 |
0.0960249178 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |