| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
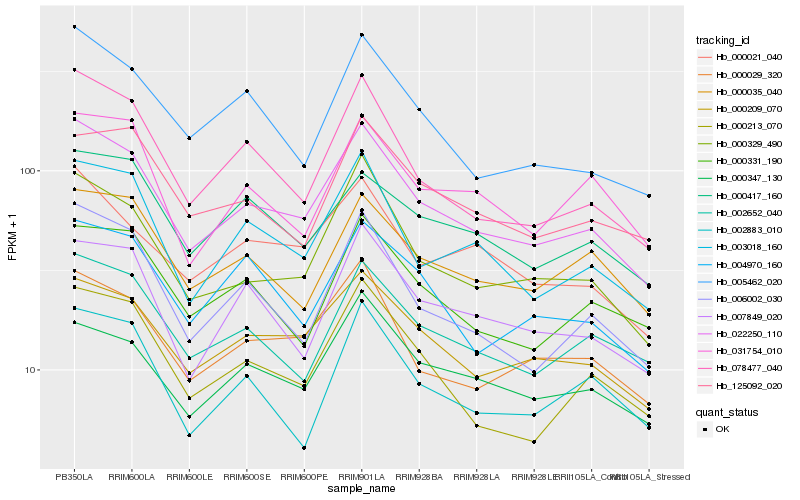
Hb_022250_110 |
0.0 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_000329_490 |
0.0649807734 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 3 |
Hb_000209_070 |
0.0649834921 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 4 |
Hb_000029_320 |
0.0715994897 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_078477_040 |
0.0718409264 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 6 |
Hb_003018_160 |
0.0727768496 |
- |
- |
PREDICTED: uncharacterized protein LOC105628876 [Jatropha curcas] |
| 7 |
Hb_006002_030 |
0.0745076353 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002652_040 |
0.0785457092 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 9 |
Hb_000213_070 |
0.079908431 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000021_040 |
0.0801613488 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 11 |
Hb_005462_020 |
0.0806390778 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 12 |
Hb_007849_020 |
0.0836945128 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 13 |
Hb_000347_130 |
0.0840985103 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 14 |
Hb_000035_040 |
0.0846190132 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 15 |
Hb_000417_160 |
0.0883842273 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 16 |
Hb_004970_160 |
0.088983256 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000331_190 |
0.0896951658 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 18 |
Hb_031754_010 |
0.0904579843 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_125092_020 |
0.0907083645 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002883_010 |
0.0909161917 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |