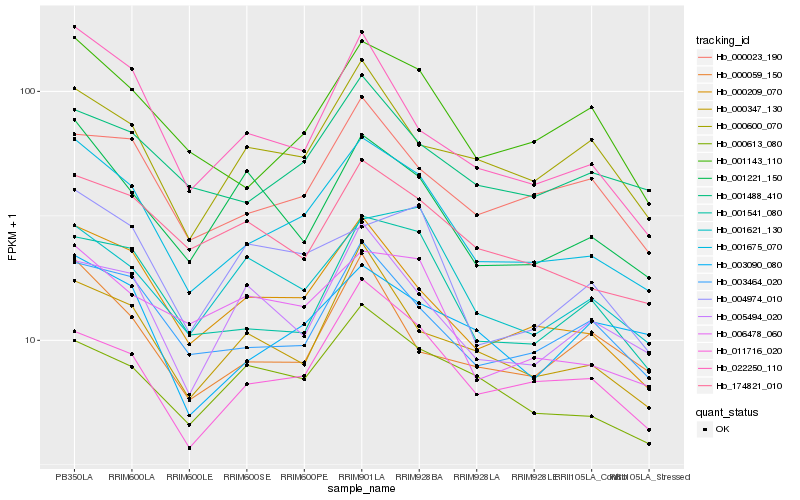
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001675_070 |
0.0 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 2 |
Hb_000209_070 |
0.0696141382 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 3 |
Hb_001541_080 |
0.0827298656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001143_110 |
0.0829913668 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 5 |
Hb_003464_020 |
0.0844286899 |
- |
- |
srpk, putative [Ricinus communis] |
| 6 |
Hb_011716_020 |
0.0847668502 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 7 |
Hb_000347_130 |
0.0893332547 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 8 |
Hb_000023_190 |
0.0906018001 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 9 |
Hb_001488_410 |
0.0930261544 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 10 |
Hb_022250_110 |
0.0931724525 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_174821_010 |
0.0941076935 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 12 |
Hb_006478_060 |
0.0943472993 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000600_070 |
0.0959037138 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 14 |
Hb_000613_080 |
0.0960965879 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 15 |
Hb_001221_150 |
0.0964399603 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 16 |
Hb_001621_130 |
0.0981256913 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003090_080 |
0.0996794745 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein B [Jatropha curcas] |
| 18 |
Hb_000059_150 |
0.1020727022 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_004974_010 |
0.1020939965 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 20 |
Hb_005494_020 |
0.1023188328 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |