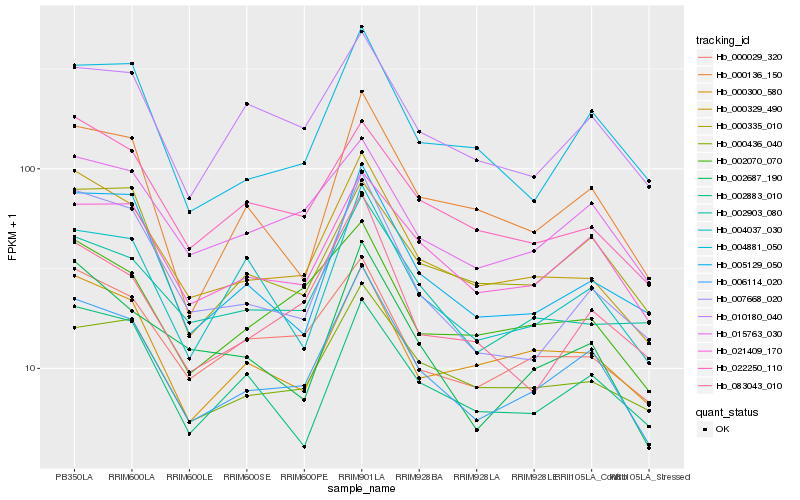
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006114_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000329_490 |
0.0734306447 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 3 |
Hb_021409_170 |
0.0750098249 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_015763_030 |
0.0852028305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004881_050 |
0.0892358812 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 6 |
Hb_010180_040 |
0.094414446 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 7 |
Hb_000335_010 |
0.0944548172 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_005129_050 |
0.0949814007 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase [Jatropha curcas] |
| 9 |
Hb_000029_320 |
0.096868385 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 10 |
Hb_000300_580 |
0.096901989 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 11 |
Hb_002070_070 |
0.0970749221 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000436_040 |
0.0978157668 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 13 |
Hb_002687_190 |
0.1002663 |
- |
- |
prolyl oligopeptidase [Cicer arietinum] |
| 14 |
Hb_000136_150 |
0.1005093642 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 15 |
Hb_004037_030 |
0.1007137796 |
- |
- |
PREDICTED: DNA repair endonuclease UVH1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007668_020 |
0.1011832998 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 17 |
Hb_002883_010 |
0.1034256538 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_022250_110 |
0.1037335657 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_083043_010 |
0.1039045552 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 20 |
Hb_002903_080 |
0.1049025318 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |