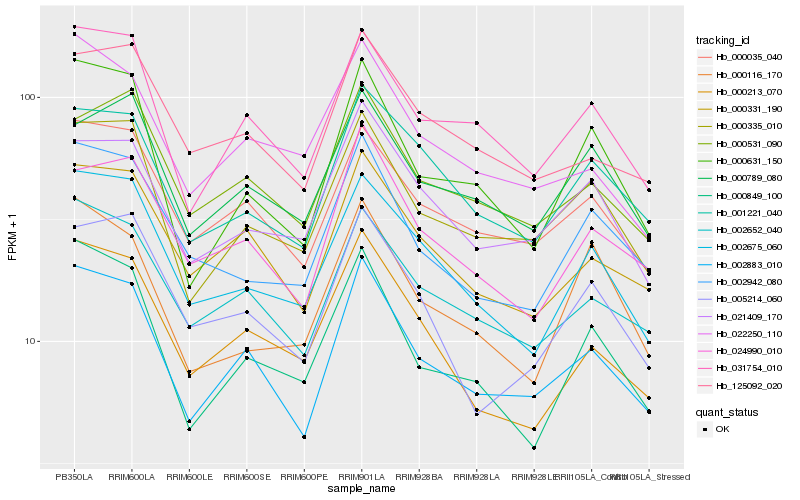
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002675_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001221_040 |
0.0722681324 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 3 |
Hb_000213_070 |
0.0727063722 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000035_040 |
0.0731943865 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 5 |
Hb_002652_040 |
0.0738531871 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 6 |
Hb_002942_080 |
0.0757519759 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 7 |
Hb_000331_190 |
0.0792659716 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 8 |
Hb_031754_010 |
0.0796751025 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000335_010 |
0.0820732779 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_005214_060 |
0.08208785 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000849_100 |
0.0830652077 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 12 |
Hb_021409_170 |
0.0841892105 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_125092_020 |
0.0865712224 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002883_010 |
0.0876134683 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_024990_010 |
0.0882635776 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 16 |
Hb_000531_090 |
0.0888172771 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 17 |
Hb_000116_170 |
0.0888981095 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 18 |
Hb_000789_080 |
0.0901519993 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_022250_110 |
0.091745477 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000631_150 |
0.0925363161 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |