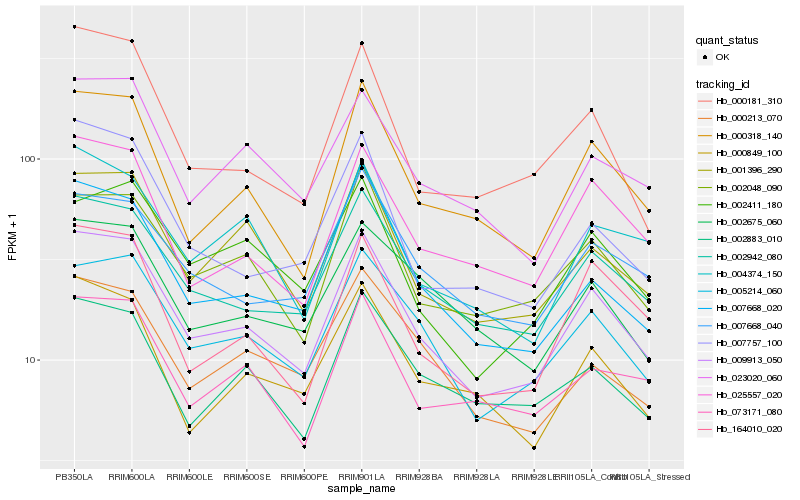
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009913_050 |
0.0 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 2 |
Hb_005214_060 |
0.064654391 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_002942_080 |
0.0647225173 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 4 |
Hb_000318_140 |
0.0744009757 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_001396_290 |
0.0780310659 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 6 |
Hb_002048_090 |
0.0797689719 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 7 |
Hb_025557_020 |
0.081413625 |
- |
- |
PREDICTED: activating signal cointegrator 1 [Jatropha curcas] |
| 8 |
Hb_007668_020 |
0.0833722385 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 9 |
Hb_023020_060 |
0.0858176941 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 10 |
Hb_164010_020 |
0.087009992 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 11 |
Hb_007668_040 |
0.0891118025 |
- |
- |
PREDICTED: uncharacterized protein LOC103409082 [Malus domestica] |
| 12 |
Hb_000181_310 |
0.0917181008 |
- |
- |
hypothetical protein JCGZ_20998 [Jatropha curcas] |
| 13 |
Hb_000849_100 |
0.0918861273 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 14 |
Hb_002411_180 |
0.0924858613 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_073171_080 |
0.093152574 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_007757_100 |
0.0940612522 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 17 |
Hb_002883_010 |
0.0942957958 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002675_060 |
0.0943470852 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004374_150 |
0.09460288 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000213_070 |
0.0952501273 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |