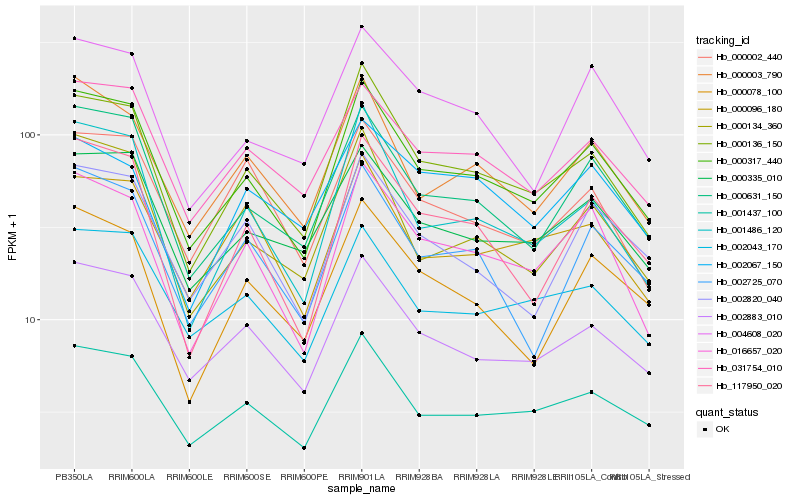
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016657_020 |
0.0 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 2 |
Hb_000317_440 |
0.0695716326 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000136_150 |
0.0774965974 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 4 |
Hb_117950_020 |
0.094431971 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 5 |
Hb_000003_790 |
0.0990774622 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 6 |
Hb_000631_150 |
0.099182578 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 7 |
Hb_001486_120 |
0.0995227007 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 8 |
Hb_002883_010 |
0.1031434353 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001437_100 |
0.1032450939 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 10 |
Hb_004608_020 |
0.1038570494 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 11 |
Hb_000096_180 |
0.1072350891 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g67570, chloroplastic-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000002_440 |
0.1090789838 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 13 |
Hb_031754_010 |
0.1102745072 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_000078_100 |
0.1121546597 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_002820_040 |
0.1156951795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000335_010 |
0.1156983315 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_002067_150 |
0.1161264257 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 18 |
Hb_002043_170 |
0.1165059019 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 19 |
Hb_000134_360 |
0.1179718422 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 20 |
Hb_002725_070 |
0.1186921421 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |