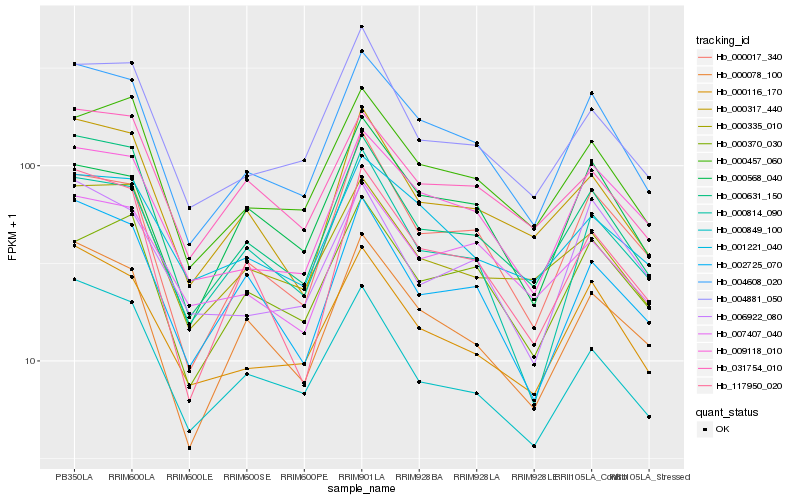
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004608_020 |
0.0 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 2 |
Hb_009118_010 |
0.0654483929 |
- |
- |
methionine aminopeptidase, putative [Ricinus communis] |
| 3 |
Hb_000631_150 |
0.0657544887 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 4 |
Hb_000116_170 |
0.0664896781 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 5 |
Hb_000457_060 |
0.0701854281 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_000017_340 |
0.0749400545 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 7 |
Hb_000078_100 |
0.0751201084 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000317_440 |
0.0842588619 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004881_050 |
0.0873307955 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 10 |
Hb_000814_090 |
0.0880813863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000370_030 |
0.0901838209 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 12 |
Hb_000568_040 |
0.0904913232 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_117950_020 |
0.0927015949 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 14 |
Hb_001221_040 |
0.0929193273 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 15 |
Hb_002725_070 |
0.0930392384 |
- |
- |
hypothetical protein PRUPE_ppa005949mg [Prunus persica] |
| 16 |
Hb_007407_040 |
0.0949456562 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_031754_010 |
0.0967584727 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_000335_010 |
0.0976446347 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000849_100 |
0.0989668401 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 20 |
Hb_006922_080 |
0.0997510088 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT3-like [Glycine max] |