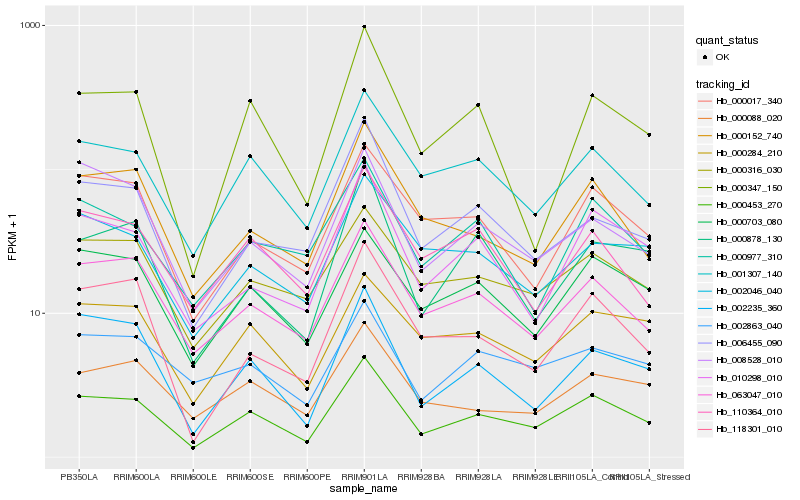
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_270 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 2 |
Hb_001307_140 |
0.0940561304 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Jatropha curcas] |
| 3 |
Hb_063047_010 |
0.0988940195 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 4 |
Hb_000703_080 |
0.1069580205 |
- |
- |
guanine nucleotide exchange factor P532, putative [Ricinus communis] |
| 5 |
Hb_002235_360 |
0.1088573823 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 6 |
Hb_010298_010 |
0.1109268322 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 7 |
Hb_000152_740 |
0.1169832741 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 8 |
Hb_000316_030 |
0.117673759 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000347_150 |
0.1187538055 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 10 |
Hb_000017_340 |
0.1197953842 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 11 |
Hb_000088_020 |
0.1201436171 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 12 |
Hb_000284_210 |
0.1205733396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006455_090 |
0.1228455696 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 14 |
Hb_118301_010 |
0.1234693225 |
- |
- |
Endoribonuclease Dicer-1-like protein [Morus notabilis] |
| 15 |
Hb_000977_310 |
0.1239334905 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 isoform X4 [Jatropha curcas] |
| 16 |
Hb_110364_010 |
0.1243081585 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 17 |
Hb_008528_010 |
0.1249591403 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 18 |
Hb_000878_130 |
0.1265175558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002863_040 |
0.1277877058 |
- |
- |
PREDICTED: uncharacterized protein LOC105630268 [Jatropha curcas] |
| 20 |
Hb_002046_040 |
0.1280141863 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |