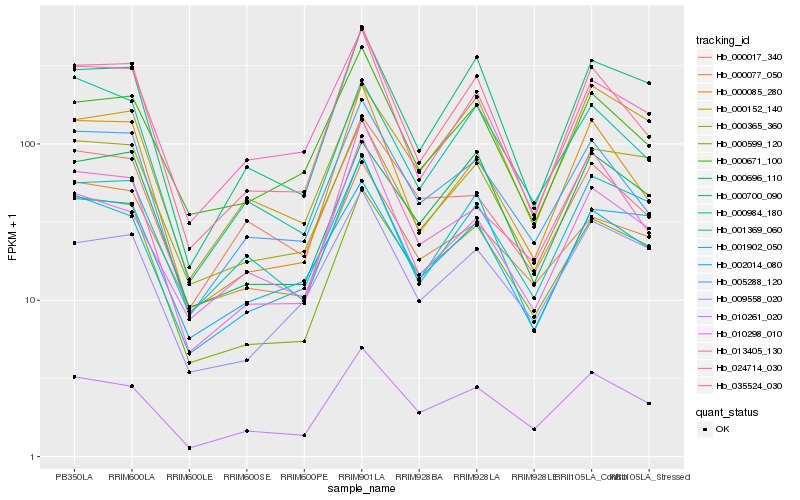
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010261_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_17258 [Jatropha curcas] |
| 2 |
Hb_005288_120 |
0.0759377442 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 3 |
Hb_013405_130 |
0.0886146376 |
- |
- |
hypothetical protein CICLE_v10032221mg [Citrus clementina] |
| 4 |
Hb_010298_010 |
0.0888696371 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 5 |
Hb_009558_020 |
0.0899796281 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000671_100 |
0.0907105741 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |
| 7 |
Hb_000700_090 |
0.0913919628 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_035524_030 |
0.0916883864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000599_120 |
0.0938598404 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT34 [Jatropha curcas] |
| 10 |
Hb_002014_080 |
0.0939778076 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000017_340 |
0.0981450206 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 12 |
Hb_001902_050 |
0.0999317605 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |
| 13 |
Hb_000365_360 |
0.1010522631 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001369_060 |
0.1041216872 |
- |
- |
PREDICTED: uncharacterized Rho GTPase-activating protein At5g61530 [Jatropha curcas] |
| 15 |
Hb_000696_110 |
0.1049209246 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000984_180 |
0.1053990266 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000152_140 |
0.1064463615 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000077_050 |
0.1065852555 |
- |
- |
PREDICTED: elongation factor G-1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_024714_030 |
0.1072923856 |
- |
- |
PREDICTED: sm-like protein LSM4 [Jatropha curcas] |
| 20 |
Hb_000085_280 |
0.1085038911 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |