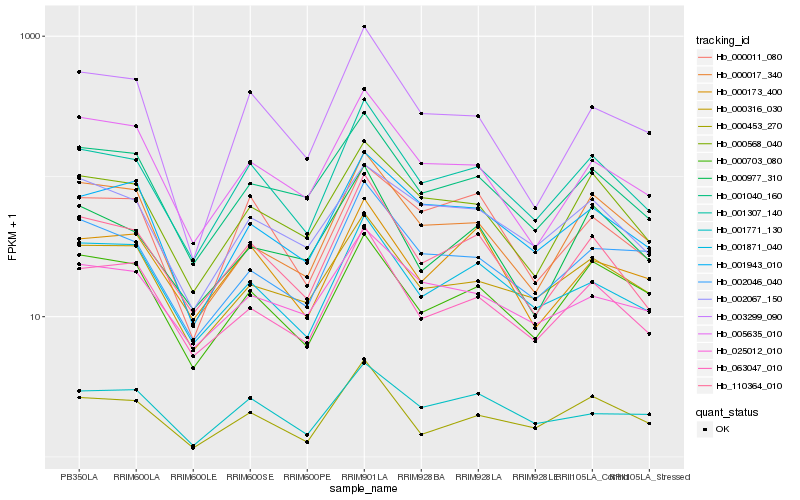
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001307_140 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Jatropha curcas] |
| 2 |
Hb_110364_010 |
0.0608484189 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 3 |
Hb_063047_010 |
0.0813888342 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 4 |
Hb_001040_160 |
0.083893735 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 5 |
Hb_000568_040 |
0.0845208923 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2 [Jatropha curcas] |
| 6 |
Hb_000453_270 |
0.0940561304 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 7 |
Hb_001943_010 |
0.0949953803 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 8 |
Hb_001871_040 |
0.0961727599 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000316_030 |
0.0974096635 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_001771_130 |
0.0997482692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g27610 [Jatropha curcas] |
| 11 |
Hb_002046_040 |
0.1004561989 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 12 |
Hb_000703_080 |
0.1020141547 |
- |
- |
guanine nucleotide exchange factor P532, putative [Ricinus communis] |
| 13 |
Hb_025012_010 |
0.1021994794 |
- |
- |
PREDICTED: importin-4 [Vitis vinifera] |
| 14 |
Hb_000017_340 |
0.1034691872 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 15 |
Hb_005635_010 |
0.1036856554 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_000977_310 |
0.1039122624 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 isoform X4 [Jatropha curcas] |
| 17 |
Hb_003299_090 |
0.1049515543 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 18 |
Hb_000011_080 |
0.1069272336 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 19 |
Hb_002067_150 |
0.1071198592 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 20 |
Hb_000173_400 |
0.1072397325 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |