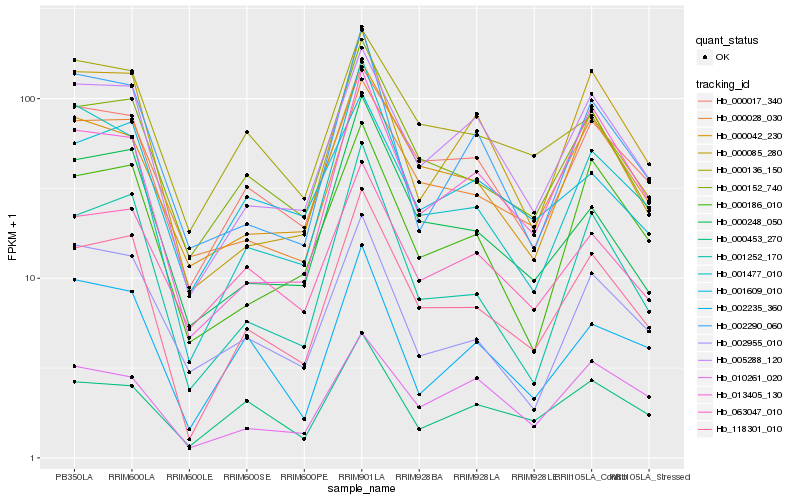
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118301_010 |
0.0 |
- |
- |
Endoribonuclease Dicer-1-like protein [Morus notabilis] |
| 2 |
Hb_000152_740 |
0.0823745807 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 3 |
Hb_000017_340 |
0.095852542 |
- |
- |
PREDICTED: GMP synthase [glutamine-hydrolyzing]-like [Phoenix dactylifera] |
| 4 |
Hb_005288_120 |
0.0964698716 |
- |
- |
PREDICTED: uncharacterized protein LOC105647751 [Jatropha curcas] |
| 5 |
Hb_001252_170 |
0.0971292922 |
desease resistance |
Gene Name: Exostosin |
hypothetical protein PRUPE_ppa001357mg [Prunus persica] |
| 6 |
Hb_001477_010 |
0.1032124907 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 13-like [Cicer arietinum] |
| 7 |
Hb_000028_030 |
0.1077112343 |
- |
- |
amidase, putative [Ricinus communis] |
| 8 |
Hb_000248_050 |
0.1080218004 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000042_230 |
0.1128360071 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 [Jatropha curcas] |
| 10 |
Hb_000186_010 |
0.1148329335 |
- |
- |
PREDICTED: EH domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_013405_130 |
0.115487536 |
- |
- |
hypothetical protein CICLE_v10032221mg [Citrus clementina] |
| 12 |
Hb_063047_010 |
0.1190713777 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 13 |
Hb_000453_270 |
0.1234693225 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 14 |
Hb_000085_280 |
0.1248882687 |
- |
- |
PREDICTED: beta-(1,2)-xylosyltransferase [Jatropha curcas] |
| 15 |
Hb_001609_010 |
0.1299687473 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative inactive disease susceptibility protein LOV1 [Jatropha curcas] |
| 16 |
Hb_002235_360 |
0.1310719191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 17 |
Hb_000136_150 |
0.1312348291 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 18 |
Hb_002290_060 |
0.1316431537 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 19 |
Hb_010261_020 |
0.1319045392 |
- |
- |
hypothetical protein JCGZ_17258 [Jatropha curcas] |
| 20 |
Hb_002955_010 |
0.1322379504 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |