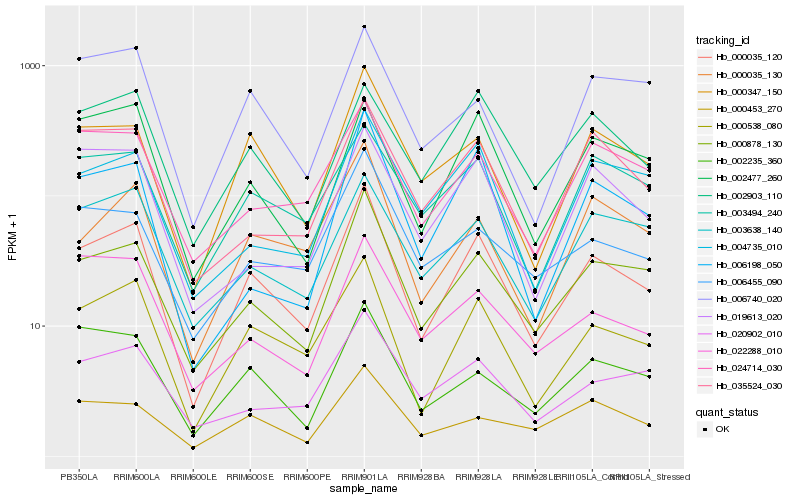
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_120 |
0.0 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 2 |
Hb_000538_080 |
0.077839995 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_000878_130 |
0.0992570148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000347_150 |
0.1188264764 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 5 |
Hb_000035_130 |
0.1274503817 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 6 |
Hb_002235_360 |
0.140236557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 7 |
Hb_002477_260 |
0.1408532504 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 8 |
Hb_020902_010 |
0.1408700906 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 9 |
Hb_006455_090 |
0.1414807093 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 10 |
Hb_024714_030 |
0.142460296 |
- |
- |
PREDICTED: sm-like protein LSM4 [Jatropha curcas] |
| 11 |
Hb_003494_240 |
0.1430625755 |
- |
- |
PREDICTED: uncharacterized protein LOC105644075 [Jatropha curcas] |
| 12 |
Hb_006198_050 |
0.143222775 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_003638_140 |
0.1432295836 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_004735_010 |
0.1456038436 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 15 |
Hb_002903_110 |
0.1459801683 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |
| 16 |
Hb_035524_030 |
0.1460245753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000453_270 |
0.1503611153 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 18 |
Hb_006740_020 |
0.1511956952 |
- |
- |
PREDICTED: bis(5'-adenosyl)-triphosphatase [Jatropha curcas] |
| 19 |
Hb_022288_010 |
0.1523711954 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 20 |
Hb_019613_020 |
0.1527141965 |
- |
- |
phospholipase C, putative [Ricinus communis] |