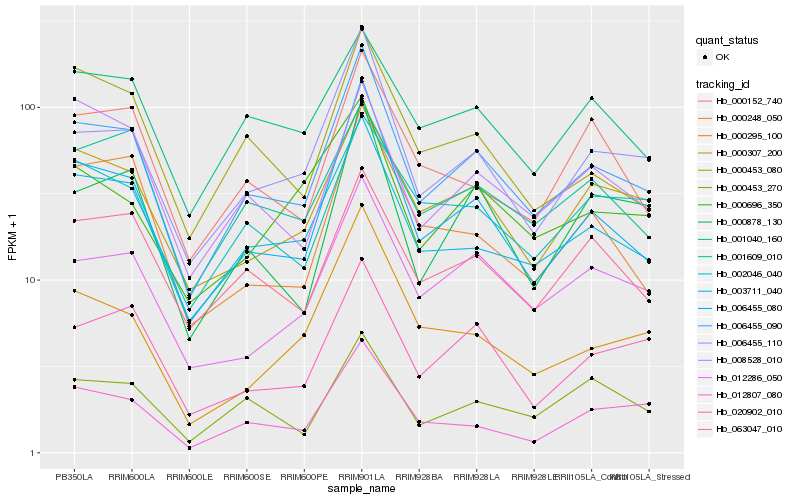
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006455_090 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 2 |
Hb_006455_080 |
0.0654894189 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 3 |
Hb_006455_110 |
0.0787941186 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003711_040 |
0.082885183 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_012286_050 |
0.1024418987 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 6 |
Hb_000307_200 |
0.1052772854 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 7 |
Hb_020902_010 |
0.1127329072 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 8 |
Hb_063047_010 |
0.1132287623 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 9 |
Hb_000878_130 |
0.1136761961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012807_080 |
0.1213595824 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 11 |
Hb_000453_080 |
0.1222014017 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000453_270 |
0.1228455696 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 13 |
Hb_001609_010 |
0.1232168272 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative inactive disease susceptibility protein LOV1 [Jatropha curcas] |
| 14 |
Hb_002046_040 |
0.1242177097 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 15 |
Hb_000295_100 |
0.1244561151 |
- |
- |
- |
| 16 |
Hb_000248_050 |
0.1248159635 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000696_350 |
0.126971822 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_001040_160 |
0.1273689481 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 19 |
Hb_000152_740 |
0.1274108867 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 20 |
Hb_008528_010 |
0.1282331665 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |