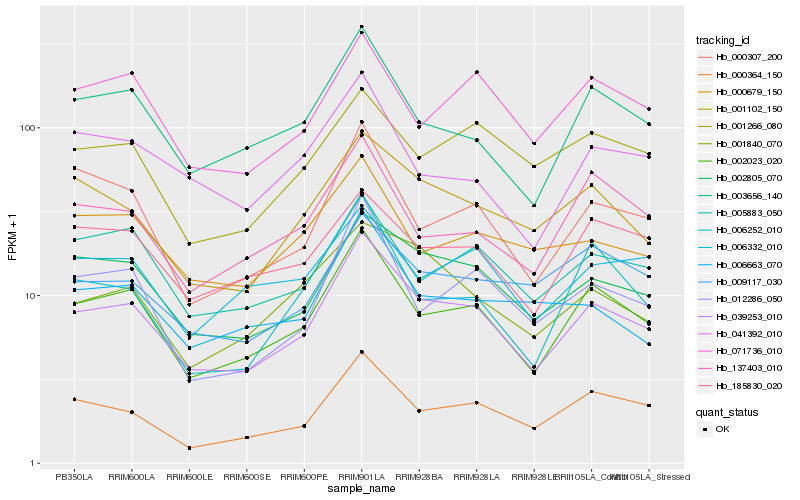
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039253_010 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002023_020 |
0.0604638509 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 3 |
Hb_002805_070 |
0.0847858437 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000364_150 |
0.094162913 |
- |
- |
- |
| 5 |
Hb_003656_140 |
0.0985707675 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001102_150 |
0.1050866556 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_012286_050 |
0.1084848908 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 8 |
Hb_000307_200 |
0.1099829092 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 9 |
Hb_005883_050 |
0.1140209508 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 10 |
Hb_071736_010 |
0.114955918 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_001840_070 |
0.1159513473 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_137403_010 |
0.1166510705 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 13 |
Hb_000679_150 |
0.1198761497 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_006663_070 |
0.1207495304 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 15 |
Hb_006252_010 |
0.1212895079 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_041392_010 |
0.1223631165 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 17 |
Hb_185830_020 |
0.1226787342 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_009117_030 |
0.1229076407 |
- |
- |
PREDICTED: histidine protein methyltransferase 1 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_006332_010 |
0.1240197043 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 isoform X1 [Vitis vinifera] |
| 20 |
Hb_001266_080 |
0.1242258949 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit gamma-like [Jatropha curcas] |