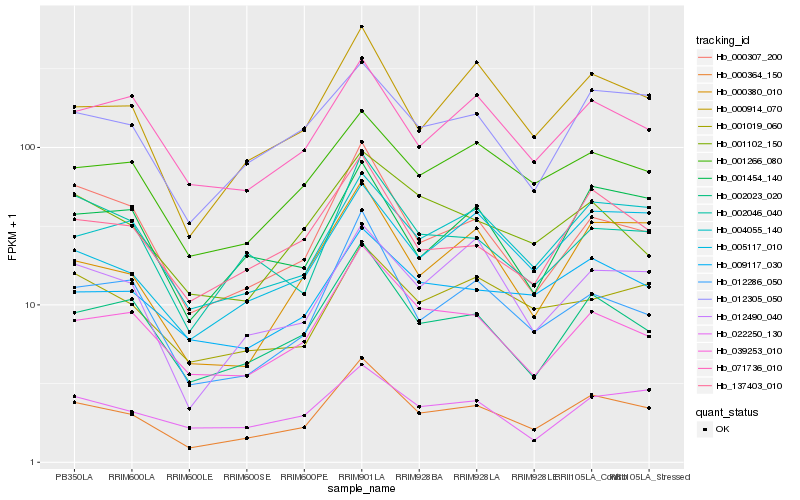
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000364_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_002023_020 |
0.086709151 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 3 |
Hb_000914_070 |
0.0905609474 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 8 [Jatropha curcas] |
| 4 |
Hb_137403_010 |
0.0912431548 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 5 |
Hb_039253_010 |
0.094162913 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004055_140 |
0.0977151015 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 7 |
Hb_009117_030 |
0.099557113 |
- |
- |
PREDICTED: histidine protein methyltransferase 1 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_001266_080 |
0.0998550733 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit gamma-like [Jatropha curcas] |
| 9 |
Hb_000307_200 |
0.0999101327 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 10 |
Hb_012305_050 |
0.0999994134 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_012286_050 |
0.1031649199 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 12 |
Hb_071736_010 |
0.1077871308 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_002046_040 |
0.1097765831 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 14 |
Hb_005117_010 |
0.1118950796 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000380_010 |
0.1128860797 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X3 [Jatropha curcas] |
| 16 |
Hb_001019_060 |
0.1134139176 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 17 |
Hb_001454_140 |
0.1140207025 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_012490_040 |
0.1154893296 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 19 |
Hb_001102_150 |
0.1156383901 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_022250_130 |
0.1157982409 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |