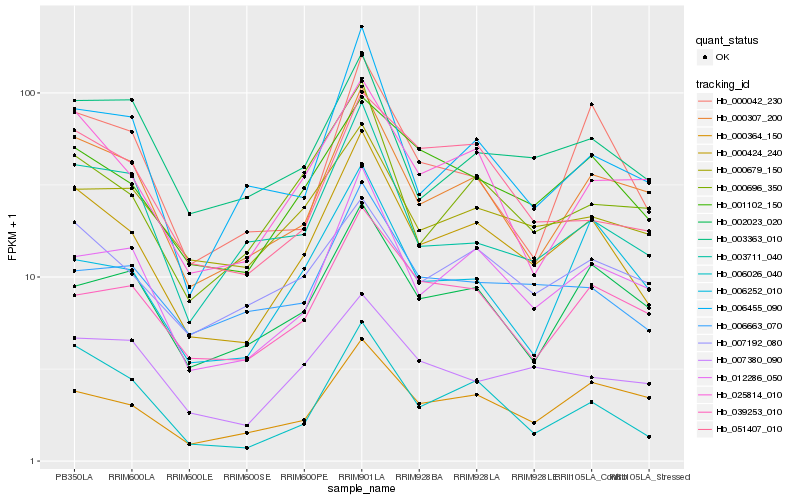
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_240 |
0.0 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 2 |
Hb_012286_050 |
0.1101683082 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 3 |
Hb_001102_150 |
0.1140010028 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000307_200 |
0.116608116 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 5 |
Hb_006026_040 |
0.1198903504 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 6 |
Hb_000696_350 |
0.1299182142 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_025814_010 |
0.1321398167 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 8 |
Hb_006663_070 |
0.1368676003 |
- |
- |
PREDICTED: uncharacterized protein LOC105637432 [Jatropha curcas] |
| 9 |
Hb_000364_150 |
0.1373909898 |
- |
- |
- |
| 10 |
Hb_007380_090 |
0.1380925343 |
- |
- |
- |
| 11 |
Hb_006455_090 |
0.1383315742 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 12 |
Hb_039253_010 |
0.1397588824 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000679_150 |
0.1399186667 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56 isoform X2 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_051407_010 |
0.1399756105 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 15 |
Hb_003363_010 |
0.1402069415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002023_020 |
0.140563997 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 17 |
Hb_000042_230 |
0.1406157141 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 [Jatropha curcas] |
| 18 |
Hb_007192_080 |
0.1406453478 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003711_040 |
0.1412120751 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_006252_010 |
0.1463628158 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 isoform X1 [Jatropha curcas] |