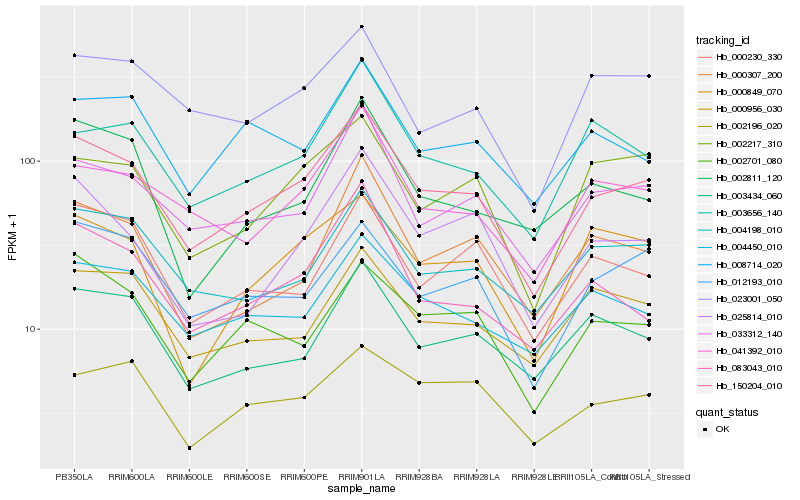
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150204_010 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 2 |
Hb_033312_140 |
0.0903304911 |
- |
- |
RNA and export factor-binding family protein [Populus trichocarpa] |
| 3 |
Hb_041392_010 |
0.0984877142 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 4 |
Hb_004198_010 |
0.102072076 |
- |
- |
Eukaryotic translation initiation factor 2 subunit beta [Theobroma cacao] |
| 5 |
Hb_004450_010 |
0.1021636767 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 6 |
Hb_083043_010 |
0.1065747774 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 7 |
Hb_003434_060 |
0.1068586566 |
- |
- |
PREDICTED: membrane-bound transcription factor site-2 protease homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_012193_010 |
0.1070076164 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_025814_010 |
0.1071855623 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 10 |
Hb_002701_080 |
0.1086359391 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 11 |
Hb_023001_050 |
0.1092750512 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 12 |
Hb_002217_310 |
0.1102919396 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 13 |
Hb_000307_200 |
0.1120318525 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 14 |
Hb_003656_140 |
0.1121092553 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008714_020 |
0.1129622535 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 16 |
Hb_000230_330 |
0.1138027386 |
- |
- |
PREDICTED: calcium-binding protein 39-like [Populus euphratica] |
| 17 |
Hb_000849_070 |
0.1140566738 |
- |
- |
PREDICTED: uncharacterized protein LOC105644190 [Jatropha curcas] |
| 18 |
Hb_000956_030 |
0.1141161865 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 19 |
Hb_002196_020 |
0.1144465928 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_002811_120 |
0.1168752024 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |