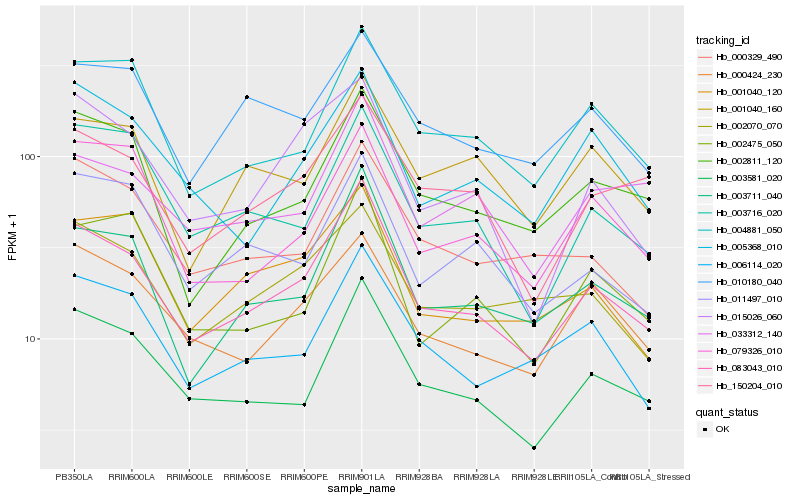
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083043_010 |
0.0 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 2 |
Hb_003711_040 |
0.0792379719 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_003581_020 |
0.0823971603 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002811_120 |
0.1005575086 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 5 |
Hb_004881_050 |
0.1007284067 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 6 |
Hb_006114_020 |
0.1039045552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_150204_010 |
0.1065747774 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 8 |
Hb_000424_230 |
0.107406455 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Glycine max] |
| 9 |
Hb_001040_120 |
0.1116657755 |
- |
- |
allantoinase, putative [Ricinus communis] |
| 10 |
Hb_002070_070 |
0.1125788835 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_005368_010 |
0.1128517867 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 12 |
Hb_001040_160 |
0.1131259397 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 13 |
Hb_000329_490 |
0.1133360246 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 14 |
Hb_015026_060 |
0.1139651143 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 15 |
Hb_010180_040 |
0.1141744067 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 16 |
Hb_003716_020 |
0.114188933 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 17 |
Hb_033312_140 |
0.1158445244 |
- |
- |
RNA and export factor-binding family protein [Populus trichocarpa] |
| 18 |
Hb_079326_010 |
0.1159600681 |
- |
- |
PREDICTED: uncharacterized protein LOC105630719 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002475_050 |
0.1166364727 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 20 |
Hb_011497_010 |
0.1171263881 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |