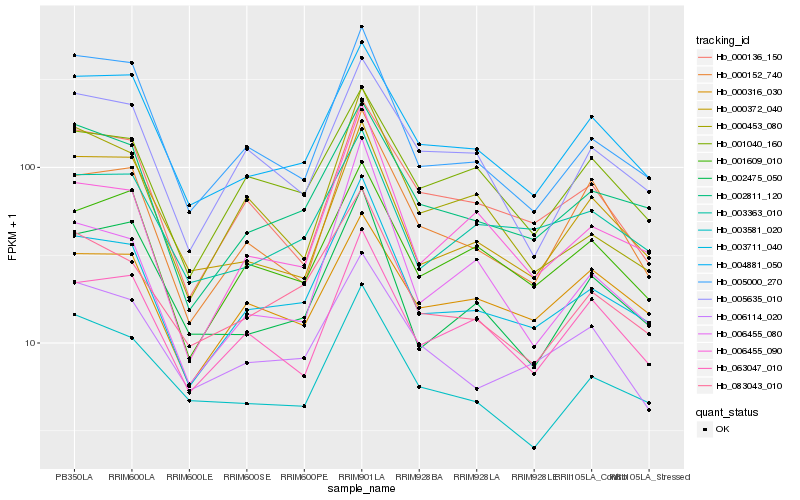
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003711_040 |
0.0 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_083043_010 |
0.0792379719 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 3 |
Hb_006455_090 |
0.082885183 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 4 |
Hb_002811_120 |
0.0842885097 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 5 |
Hb_004881_050 |
0.0957302115 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 6 |
Hb_001609_010 |
0.0984741706 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative inactive disease susceptibility protein LOV1 [Jatropha curcas] |
| 7 |
Hb_005000_270 |
0.1011763926 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_003363_010 |
0.1012550213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_063047_010 |
0.1026352733 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 10 |
Hb_001040_160 |
0.1035220898 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 11 |
Hb_000152_740 |
0.1061335454 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 12 |
Hb_006114_020 |
0.1094264685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000372_040 |
0.1105229945 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_005635_010 |
0.111253723 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 15 |
Hb_002475_050 |
0.111518799 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 16 |
Hb_000316_030 |
0.1126654288 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_003581_020 |
0.1127871688 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000453_080 |
0.1128520415 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000136_150 |
0.1134479521 |
- |
- |
hypothetical protein VITISV_038366 [Vitis vinifera] |
| 20 |
Hb_006455_080 |
0.1140505424 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |