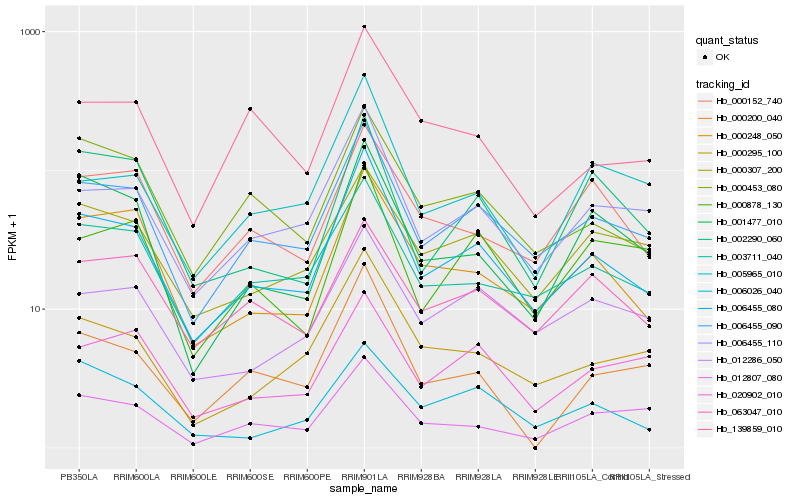
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006455_080 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_006455_090 |
0.0654894189 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 3 |
Hb_006455_110 |
0.0958485691 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000248_050 |
0.1137015938 |
- |
- |
PREDICTED: uncharacterized protein LOC105647198 isoform X3 [Jatropha curcas] |
| 5 |
Hb_003711_040 |
0.1140505424 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_000295_100 |
0.1204176559 |
- |
- |
- |
| 7 |
Hb_000453_080 |
0.1263948296 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001477_010 |
0.130326286 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 13-like [Cicer arietinum] |
| 9 |
Hb_012286_050 |
0.1349472119 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 10 |
Hb_005965_010 |
0.1363532952 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 11 |
Hb_000307_200 |
0.1385612361 |
- |
- |
PREDICTED: uncharacterized protein LOC105638529 [Jatropha curcas] |
| 12 |
Hb_000152_740 |
0.1392760733 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 13 |
Hb_012807_080 |
0.1398918051 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 14 |
Hb_139859_010 |
0.1411727788 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 15 |
Hb_000878_130 |
0.1418679108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006026_040 |
0.1460599353 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 17 |
Hb_063047_010 |
0.1463005027 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 18 |
Hb_020902_010 |
0.1470301391 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 19 |
Hb_000200_040 |
0.1473114821 |
- |
- |
- |
| 20 |
Hb_002290_060 |
0.1474905693 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |