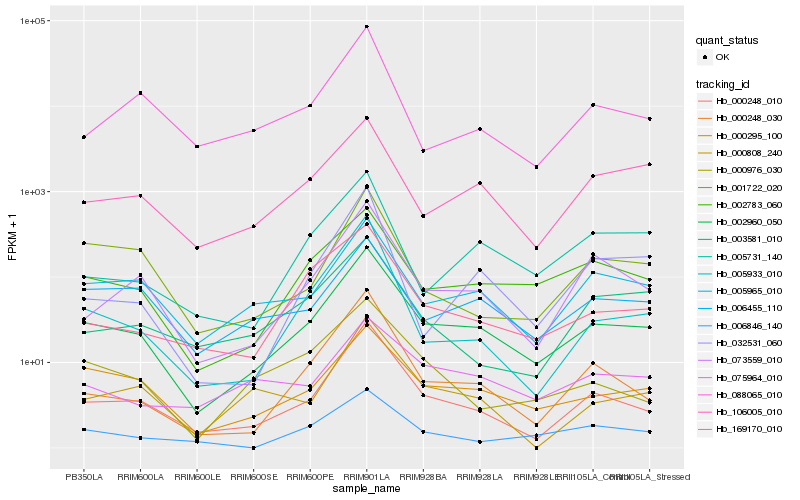
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 2 |
Hb_000248_010 |
0.1152481656 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 3 |
Hb_000295_100 |
0.127874474 |
- |
- |
- |
| 4 |
Hb_002783_060 |
0.1283809807 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 5 |
Hb_005965_010 |
0.1394517168 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 6 |
Hb_073559_010 |
0.1506757879 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 7 |
Hb_106005_010 |
0.1551193715 |
- |
- |
EG5651 [Manihot esculenta] |
| 8 |
Hb_169170_010 |
0.1572896167 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_000248_030 |
0.1616693525 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 10 |
Hb_006455_110 |
0.1616775418 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005731_140 |
0.1708457891 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 12 |
Hb_000976_030 |
0.1724760524 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like [Citrus sinensis] |
| 13 |
Hb_006846_140 |
0.1750859687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001722_020 |
0.1755801434 |
- |
- |
- |
| 15 |
Hb_088065_010 |
0.1789788009 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 16 |
Hb_000808_240 |
0.1791096349 |
- |
- |
- |
| 17 |
Hb_075964_010 |
0.1799323106 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 18 |
Hb_005933_010 |
0.1854804132 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 19 |
Hb_003581_010 |
0.1861391662 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 20 |
Hb_032531_060 |
0.1895447693 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |