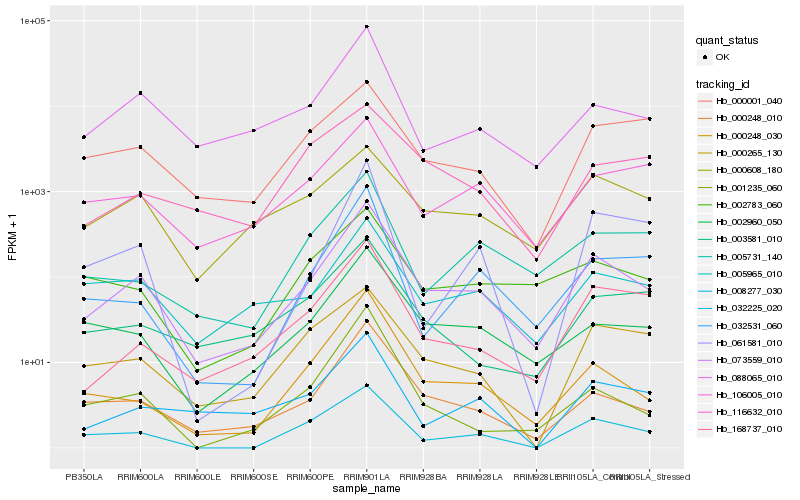
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073559_010 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 2 |
Hb_000248_010 |
0.1301616661 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 3 |
Hb_088065_010 |
0.1479674224 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 4 |
Hb_002960_050 |
0.1506757879 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 5 |
Hb_000248_030 |
0.1520026962 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 6 |
Hb_168737_010 |
0.1600451737 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_005965_010 |
0.1667463914 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 8 |
Hb_008277_030 |
0.1671306449 |
- |
- |
hypothetical protein CICLE_v10024419mg, partial [Citrus clementina] |
| 9 |
Hb_106005_010 |
0.1750378248 |
- |
- |
EG5651 [Manihot esculenta] |
| 10 |
Hb_005731_140 |
0.1771213875 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 11 |
Hb_032225_020 |
0.1787188339 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 12 |
Hb_032531_060 |
0.1794965793 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 13 |
Hb_002783_060 |
0.1825717801 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 14 |
Hb_003581_010 |
0.182880746 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 15 |
Hb_000265_130 |
0.1839387988 |
- |
- |
- |
| 16 |
Hb_000608_180 |
0.1889999668 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |
| 17 |
Hb_001235_060 |
0.1896735607 |
- |
- |
- |
| 18 |
Hb_000001_040 |
0.1955712477 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_061581_010 |
0.1995970712 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_116632_010 |
0.2056491039 |
- |
- |
unnamed protein product [Vitis vinifera] |