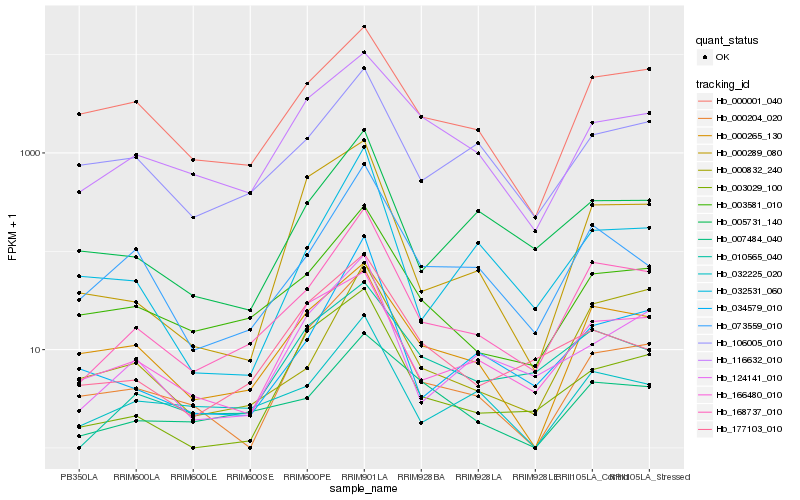
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168737_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_003581_010 |
0.1403766784 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 3 |
Hb_073559_010 |
0.1600451737 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 4 |
Hb_032225_020 |
0.1678352435 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 5 |
Hb_005731_140 |
0.1693786289 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 6 |
Hb_000265_130 |
0.1735982803 |
- |
- |
- |
| 7 |
Hb_000832_240 |
0.1749395549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_034579_010 |
0.1766073702 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 9 |
Hb_106005_010 |
0.1778808531 |
- |
- |
EG5651 [Manihot esculenta] |
| 10 |
Hb_124141_010 |
0.177989076 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 11 |
Hb_010565_040 |
0.1786147018 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 12 |
Hb_000001_040 |
0.1799005982 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_007484_040 |
0.1840682139 |
- |
- |
- |
| 14 |
Hb_116632_010 |
0.1859782638 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_032531_060 |
0.1867663004 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 16 |
Hb_166480_010 |
0.1870634443 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 17 |
Hb_000204_020 |
0.1880267232 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 18 |
Hb_177103_010 |
0.1994883786 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 19 |
Hb_000289_080 |
0.2039144569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003029_100 |
0.2068648198 |
- |
- |
- |