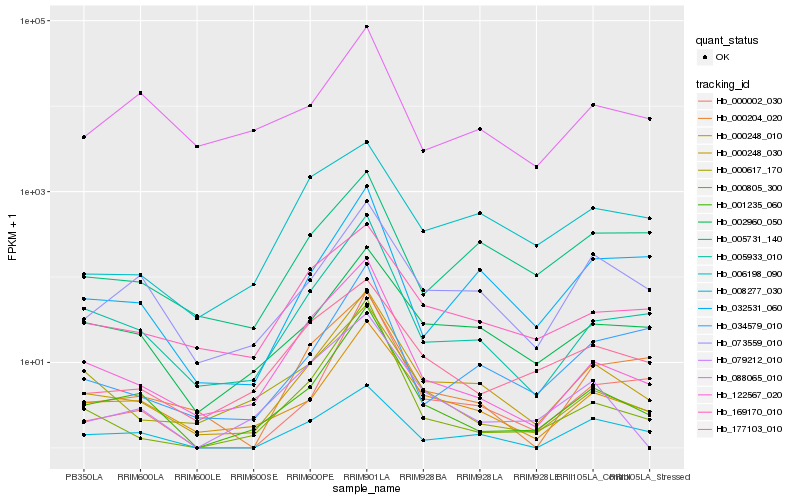
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000248_030 |
0.0 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 2 |
Hb_000248_010 |
0.1206355984 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 3 |
Hb_005933_010 |
0.1347181018 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 4 |
Hb_001235_060 |
0.1377244435 |
- |
- |
- |
| 5 |
Hb_122567_020 |
0.1512647604 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 6 |
Hb_073559_010 |
0.1520026962 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 7 |
Hb_002960_050 |
0.1616693525 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 8 |
Hb_169170_010 |
0.1690725291 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_032531_060 |
0.1691201383 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 10 |
Hb_000002_030 |
0.1788914931 |
- |
- |
- |
| 11 |
Hb_079212_010 |
0.184059994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000204_020 |
0.1867299253 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 13 |
Hb_000617_170 |
0.1869258173 |
- |
- |
- |
| 14 |
Hb_000805_300 |
0.1877483594 |
- |
- |
- |
| 15 |
Hb_177103_010 |
0.187975082 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 16 |
Hb_034579_010 |
0.1879996162 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005731_140 |
0.1956714811 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 18 |
Hb_008277_030 |
0.1971684022 |
- |
- |
hypothetical protein CICLE_v10024419mg, partial [Citrus clementina] |
| 19 |
Hb_088065_010 |
0.2040780872 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 20 |
Hb_006198_090 |
0.2047043057 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |