| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
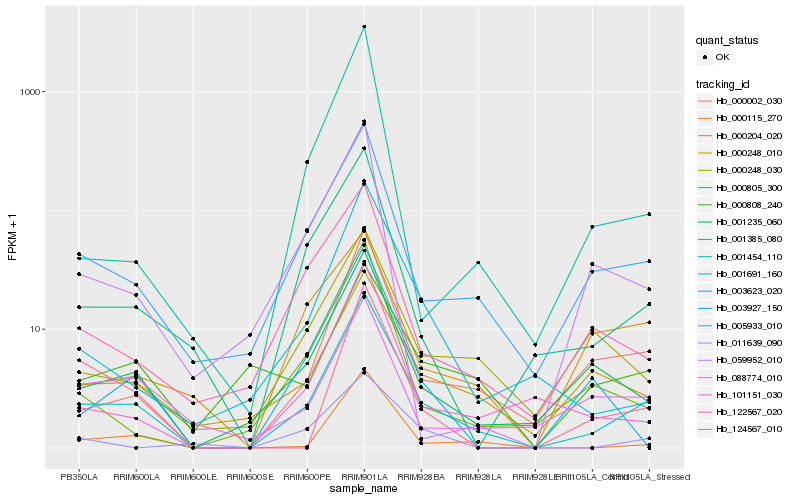
Hb_001691_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005933_010 |
0.1594527596 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 3 |
Hb_000002_030 |
0.1761653694 |
- |
- |
- |
| 4 |
Hb_001235_060 |
0.1822513777 |
- |
- |
- |
| 5 |
Hb_000115_270 |
0.1956178947 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s13290g [Populus trichocarpa] |
| 6 |
Hb_001385_080 |
0.2013853001 |
- |
- |
- |
| 7 |
Hb_101151_030 |
0.204614877 |
- |
- |
- |
| 8 |
Hb_000248_010 |
0.2050471781 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 9 |
Hb_003623_020 |
0.2169473812 |
- |
- |
- |
| 10 |
Hb_122567_020 |
0.2182418108 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 11 |
Hb_000805_300 |
0.2218515923 |
- |
- |
- |
| 12 |
Hb_000248_030 |
0.2228356904 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 13 |
Hb_124567_010 |
0.2245827512 |
- |
- |
- |
| 14 |
Hb_088774_010 |
0.2312515307 |
- |
- |
- |
| 15 |
Hb_003927_150 |
0.2330179237 |
- |
- |
- |
| 16 |
Hb_001454_110 |
0.2367787162 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 17 |
Hb_000808_240 |
0.2383951807 |
- |
- |
- |
| 18 |
Hb_000204_020 |
0.2389365191 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 19 |
Hb_059952_010 |
0.2400794877 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 20 |
Hb_011639_090 |
0.2415268381 |
- |
- |
PREDICTED: transcription factor bHLH75-like [Populus euphratica] |