| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
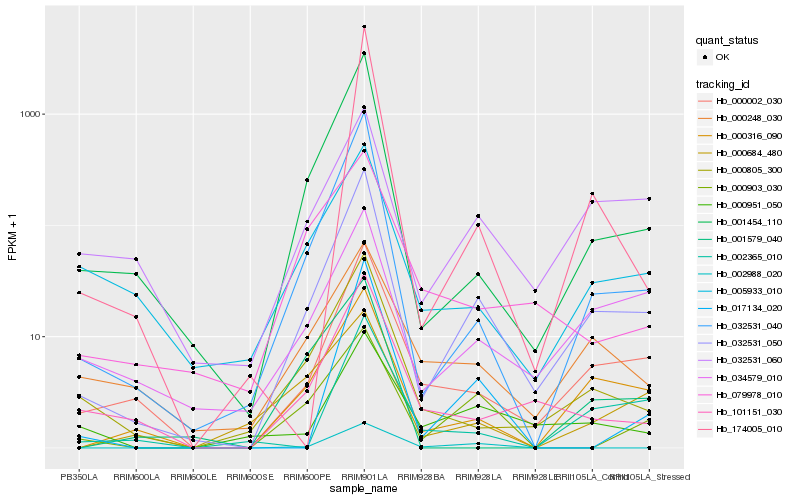
Hb_032531_050 |
0.0 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 2 |
Hb_032531_040 |
0.1450428715 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 3 |
Hb_000002_030 |
0.1534231241 |
- |
- |
- |
| 4 |
Hb_001454_110 |
0.1548244947 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 5 |
Hb_000316_090 |
0.167814294 |
- |
- |
- |
| 6 |
Hb_000805_300 |
0.185645316 |
- |
- |
- |
| 7 |
Hb_034579_010 |
0.2013753648 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 8 |
Hb_101151_030 |
0.204713702 |
- |
- |
- |
| 9 |
Hb_032531_060 |
0.2111904408 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 10 |
Hb_174005_010 |
0.2204793573 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 11 |
Hb_000903_030 |
0.2215255244 |
- |
- |
- |
| 12 |
Hb_017134_020 |
0.2235448831 |
- |
- |
- |
| 13 |
Hb_000248_030 |
0.2266660701 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 14 |
Hb_005933_010 |
0.2270262144 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 15 |
Hb_002365_010 |
0.2279507727 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 16 |
Hb_000951_050 |
0.2316125048 |
- |
- |
- |
| 17 |
Hb_001579_040 |
0.2401417218 |
- |
- |
- |
| 18 |
Hb_079978_010 |
0.2406073081 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 19 |
Hb_000684_480 |
0.2431036053 |
transcription factor |
TF Family: Orphans |
- |
| 20 |
Hb_002988_020 |
0.2473175154 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |