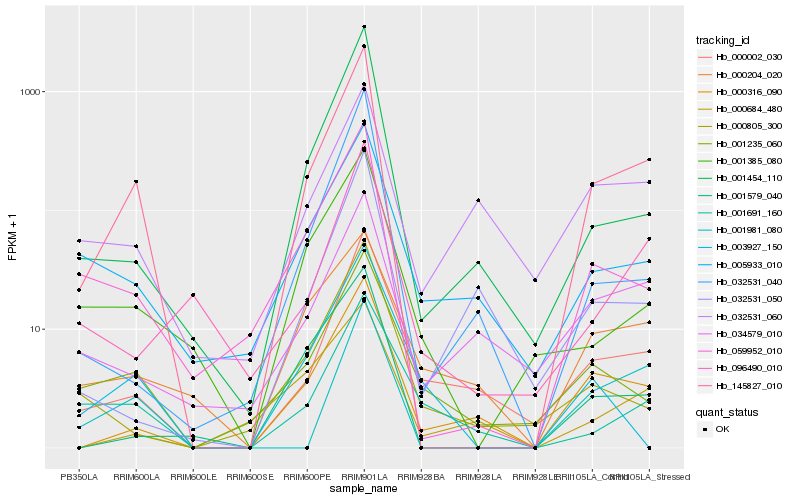
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145827_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13630g [Populus trichocarpa] |
| 2 |
Hb_059952_010 |
0.1824793174 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 3 |
Hb_000316_090 |
0.1877989993 |
- |
- |
- |
| 4 |
Hb_001579_040 |
0.1995323286 |
- |
- |
- |
| 5 |
Hb_000002_030 |
0.2035636597 |
- |
- |
- |
| 6 |
Hb_001454_110 |
0.2083530537 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 7 |
Hb_001981_080 |
0.2115693046 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 isoform X4 [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_001385_080 |
0.2267642985 |
- |
- |
- |
| 9 |
Hb_032531_040 |
0.2306108415 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 10 |
Hb_001235_060 |
0.2313942498 |
- |
- |
- |
| 11 |
Hb_096490_010 |
0.2339903638 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 12 |
Hb_005933_010 |
0.2343399724 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 13 |
Hb_034579_010 |
0.2375843351 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001691_160 |
0.2426844981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000204_020 |
0.2445912431 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 16 |
Hb_000684_480 |
0.2491178802 |
transcription factor |
TF Family: Orphans |
- |
| 17 |
Hb_032531_050 |
0.2506858124 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 18 |
Hb_000805_300 |
0.2507478142 |
- |
- |
- |
| 19 |
Hb_032531_060 |
0.2522557047 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 20 |
Hb_003927_150 |
0.2536454412 |
- |
- |
- |