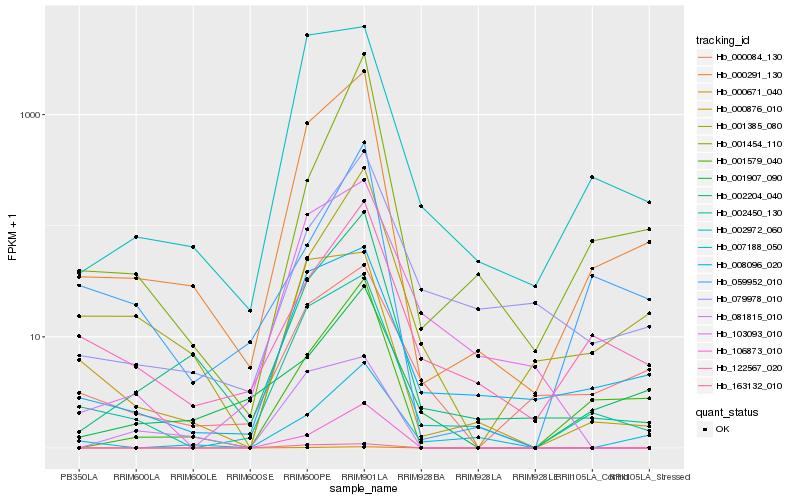
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_002450_130 |
0.1428008635 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 3 |
Hb_002204_040 |
0.1475752726 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 4 |
Hb_001579_040 |
0.163642343 |
- |
- |
- |
| 5 |
Hb_000671_040 |
0.1797111133 |
- |
- |
- |
| 6 |
Hb_008096_020 |
0.1808971421 |
- |
- |
- |
| 7 |
Hb_002972_060 |
0.196770219 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 8 |
Hb_122567_020 |
0.1989433629 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 9 |
Hb_001385_080 |
0.2011768621 |
- |
- |
- |
| 10 |
Hb_106873_010 |
0.2013243832 |
- |
- |
- |
| 11 |
Hb_007188_050 |
0.2153616764 |
- |
- |
- |
| 12 |
Hb_000876_010 |
0.2166561114 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 13 |
Hb_000084_130 |
0.2189792982 |
- |
- |
- |
| 14 |
Hb_059952_010 |
0.2199124649 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 15 |
Hb_079978_010 |
0.2206960052 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 16 |
Hb_001454_110 |
0.2242583554 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 17 |
Hb_103093_010 |
0.2250491269 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 18 |
Hb_081815_010 |
0.2284349263 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 19 |
Hb_163132_010 |
0.229378765 |
- |
- |
- |
| 20 |
Hb_001907_090 |
0.2309024979 |
- |
- |
- |