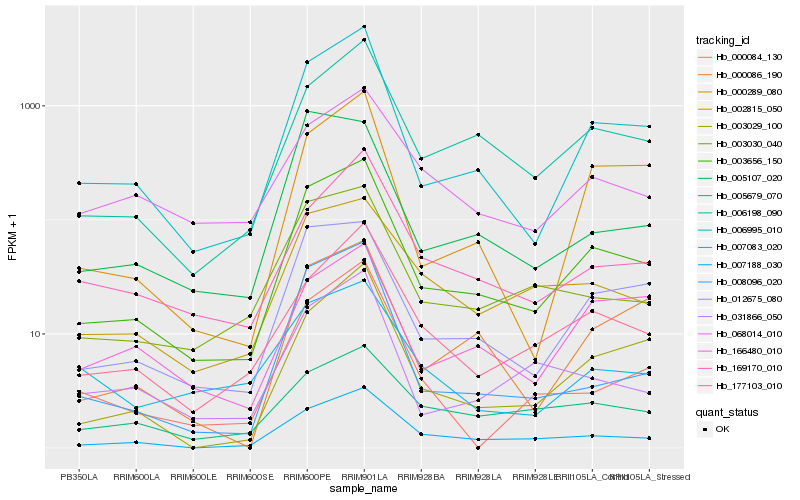
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003656_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_006995_010 |
0.0769452578 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 3 |
Hb_007188_030 |
0.1124773665 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 4 |
Hb_012675_080 |
0.1145235248 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 5 |
Hb_006198_090 |
0.1166168014 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 6 |
Hb_003029_100 |
0.136813489 |
- |
- |
- |
| 7 |
Hb_003030_040 |
0.1378603456 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 8 |
Hb_177103_010 |
0.1396070568 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 9 |
Hb_002815_050 |
0.1461284274 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 10 |
Hb_000289_080 |
0.1521082796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000086_190 |
0.1536872501 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 12 |
Hb_169170_010 |
0.1536913137 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 13 |
Hb_008096_020 |
0.1546278501 |
- |
- |
- |
| 14 |
Hb_031866_050 |
0.1576540906 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 15 |
Hb_005107_020 |
0.1686306812 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 16 |
Hb_000084_130 |
0.1717224113 |
- |
- |
- |
| 17 |
Hb_005679_070 |
0.171918675 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 18 |
Hb_068014_010 |
0.1738082671 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_166480_010 |
0.175013141 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 20 |
Hb_007083_020 |
0.17915531 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |