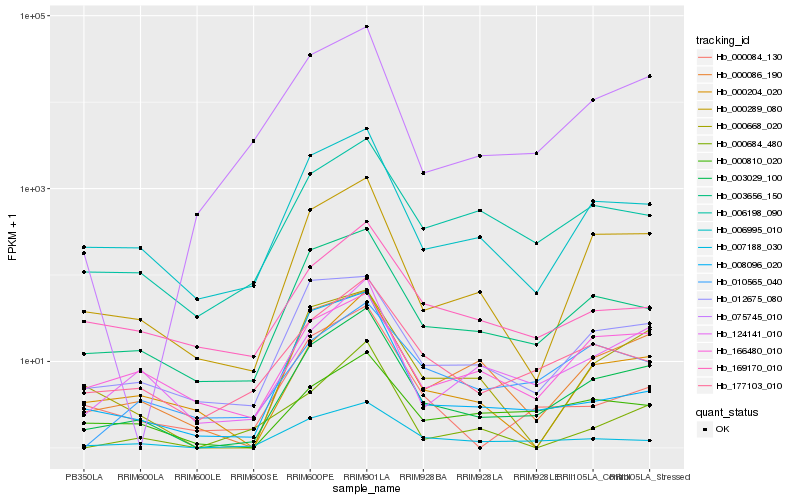
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003029_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000289_080 |
0.1226372363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006995_010 |
0.1273339436 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 4 |
Hb_003656_150 |
0.136813489 |
- |
- |
- |
| 5 |
Hb_000086_190 |
0.155139561 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 6 |
Hb_124141_010 |
0.1564227096 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 7 |
Hb_006198_090 |
0.1580344891 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 8 |
Hb_075745_010 |
0.160152905 |
- |
- |
hypothetical protein EUGRSUZ_D00579 [Eucalyptus grandis] |
| 9 |
Hb_000204_020 |
0.1609149751 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 10 |
Hb_177103_010 |
0.1609336308 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 11 |
Hb_007188_030 |
0.1624615207 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 12 |
Hb_000668_020 |
0.1713641751 |
- |
- |
- |
| 13 |
Hb_000084_130 |
0.1843527024 |
- |
- |
- |
| 14 |
Hb_012675_080 |
0.1845992304 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 15 |
Hb_169170_010 |
0.1874690586 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_166480_010 |
0.1893201996 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 17 |
Hb_000810_020 |
0.1978639959 |
- |
- |
- |
| 18 |
Hb_010565_040 |
0.1993263443 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |
| 19 |
Hb_000684_480 |
0.2044215303 |
transcription factor |
TF Family: Orphans |
- |
| 20 |
Hb_008096_020 |
0.2044240028 |
- |
- |
- |