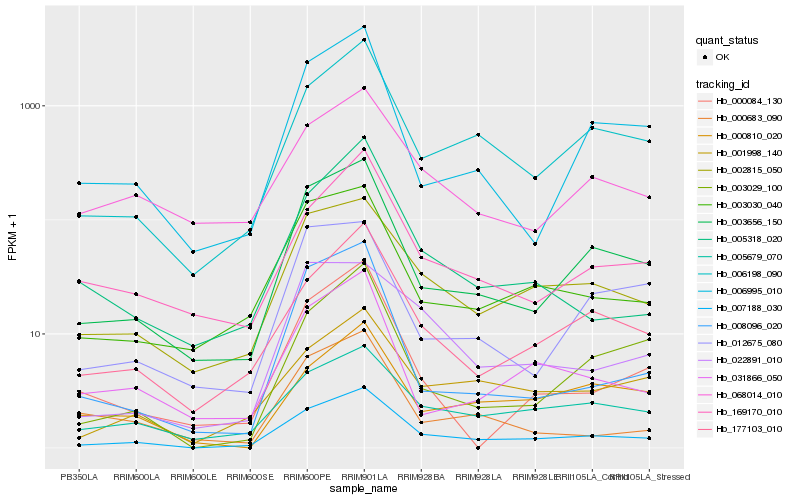
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007188_030 |
0.0 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 2 |
Hb_003656_150 |
0.1124773665 |
- |
- |
- |
| 3 |
Hb_002815_050 |
0.1156001634 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 4 |
Hb_006198_090 |
0.1197292189 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 5 |
Hb_177103_010 |
0.1250127417 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 6 |
Hb_003030_040 |
0.1366882899 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 7 |
Hb_005679_070 |
0.1414771573 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 8 |
Hb_001998_140 |
0.1433758543 |
- |
- |
- |
| 9 |
Hb_169170_010 |
0.1527598322 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 10 |
Hb_006995_010 |
0.1609490343 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 11 |
Hb_003029_100 |
0.1624615207 |
- |
- |
- |
| 12 |
Hb_031866_050 |
0.1637416142 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 13 |
Hb_068014_010 |
0.1707939299 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 14 |
Hb_005318_020 |
0.1745101193 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_022891_010 |
0.1780132873 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 16 |
Hb_000810_020 |
0.1817087894 |
- |
- |
- |
| 17 |
Hb_008096_020 |
0.1864401369 |
- |
- |
- |
| 18 |
Hb_000084_130 |
0.1877805265 |
- |
- |
- |
| 19 |
Hb_000683_090 |
0.1892815689 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 20 |
Hb_012675_080 |
0.1893539604 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |