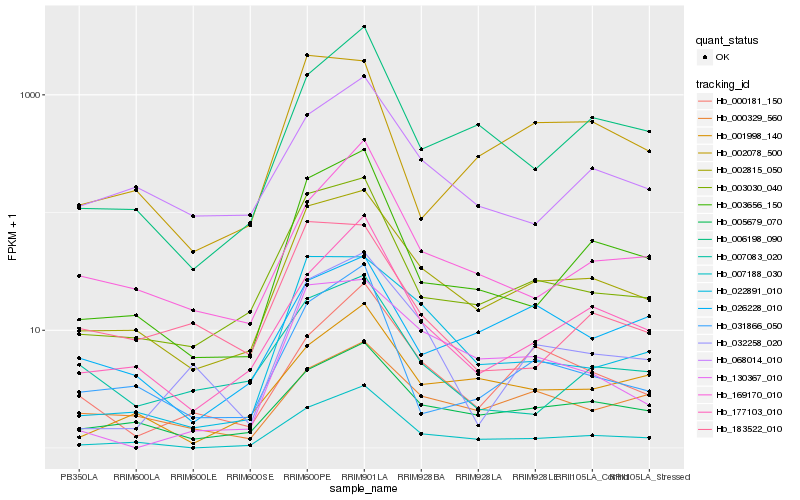
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002815_050 |
0.0 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 2 |
Hb_005679_070 |
0.0957089002 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 3 |
Hb_003030_040 |
0.1066698312 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 4 |
Hb_007188_030 |
0.1156001634 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 5 |
Hb_003656_150 |
0.1461284274 |
- |
- |
- |
| 6 |
Hb_022891_010 |
0.1461287552 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 7 |
Hb_068014_010 |
0.1468060127 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 8 |
Hb_177103_010 |
0.1602068068 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 9 |
Hb_000329_560 |
0.1662130953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006198_090 |
0.1673524918 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 11 |
Hb_001998_140 |
0.1680055871 |
- |
- |
- |
| 12 |
Hb_130367_010 |
0.1701094197 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 13 |
Hb_002078_500 |
0.1712552034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_032258_020 |
0.1733350717 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Malus domestica] |
| 15 |
Hb_000181_150 |
0.1766511957 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 16 |
Hb_031866_050 |
0.1775633647 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 17 |
Hb_007083_020 |
0.1782033026 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 18 |
Hb_026228_010 |
0.1789182726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_183522_010 |
0.1804960229 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 20 |
Hb_169170_010 |
0.1822296972 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |