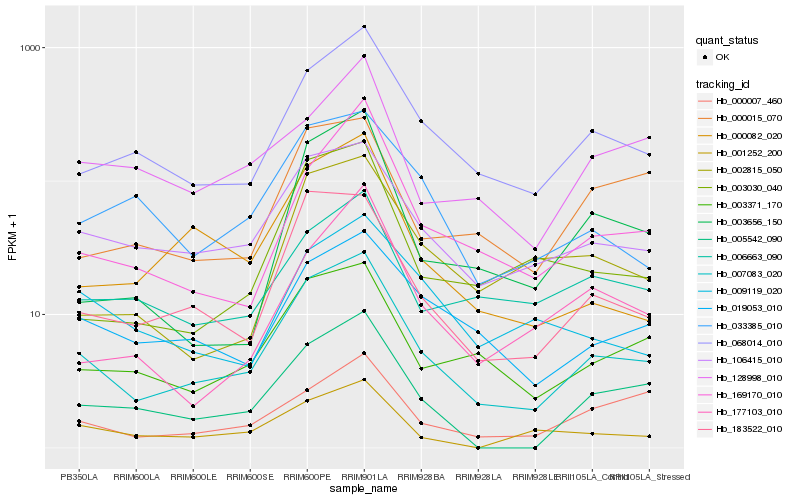
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007083_020 |
0.0 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 2 |
Hb_183522_010 |
0.1249472777 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 3 |
Hb_068014_010 |
0.1254229843 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_106415_010 |
0.1288382392 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 5 |
Hb_003371_170 |
0.1462084419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_019053_010 |
0.1557941862 |
- |
- |
- |
| 7 |
Hb_005542_090 |
0.1560417992 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 8 |
Hb_169170_010 |
0.1634144481 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_003030_040 |
0.1678214015 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 10 |
Hb_033385_010 |
0.1684185152 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 11 |
Hb_000082_020 |
0.1696047053 |
- |
- |
- |
| 12 |
Hb_006663_090 |
0.1759735346 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 13 |
Hb_001252_200 |
0.1759907193 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 14 |
Hb_002815_050 |
0.1782033026 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 15 |
Hb_000007_460 |
0.179093638 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 16 |
Hb_003656_150 |
0.17915531 |
- |
- |
- |
| 17 |
Hb_000015_070 |
0.1805961614 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 18 |
Hb_009119_020 |
0.1855621908 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 19 |
Hb_128998_010 |
0.1875833046 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 20 |
Hb_177103_010 |
0.1887180496 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |