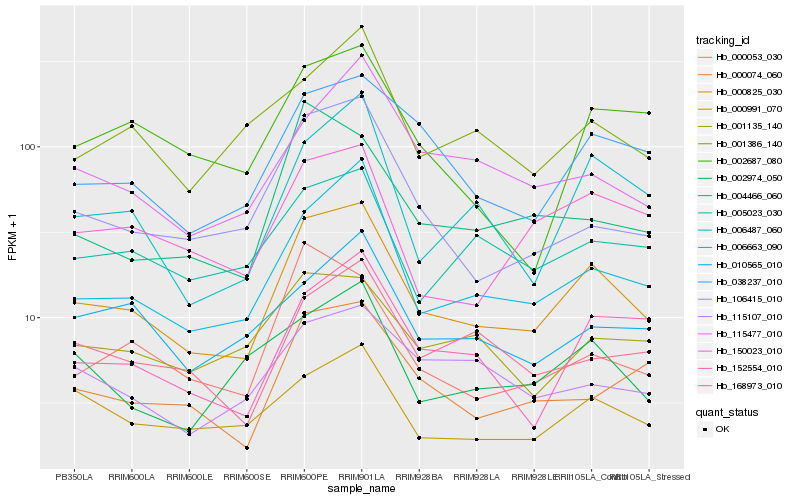
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000825_030 |
0.0 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_006663_090 |
0.10901088 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 3 |
Hb_038237_010 |
0.1254674703 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 4 |
Hb_006487_060 |
0.12859841 |
- |
- |
Defective in cullin neddylation protein, putative [Ricinus communis] |
| 5 |
Hb_106415_010 |
0.1304176914 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 6 |
Hb_000991_070 |
0.1325207482 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 7 |
Hb_152554_010 |
0.1372318023 |
- |
- |
hypothetical protein B456_008G095800, partial [Gossypium raimondii] |
| 8 |
Hb_150023_010 |
0.1402067299 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 9 |
Hb_000074_060 |
0.1404056804 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 10 |
Hb_168973_010 |
0.143381968 |
- |
- |
- |
| 11 |
Hb_005023_030 |
0.1447429302 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 12 |
Hb_001386_140 |
0.1451886615 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 13 |
Hb_115477_010 |
0.1462696245 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 14 |
Hb_002974_050 |
0.1473007222 |
- |
- |
- |
| 15 |
Hb_004466_060 |
0.1485490224 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |
| 16 |
Hb_001135_140 |
0.1488153127 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 17 |
Hb_010565_010 |
0.149039535 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000053_030 |
0.1499830196 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 19 |
Hb_002687_080 |
0.1514864838 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 20 |
Hb_115107_010 |
0.1528289065 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |