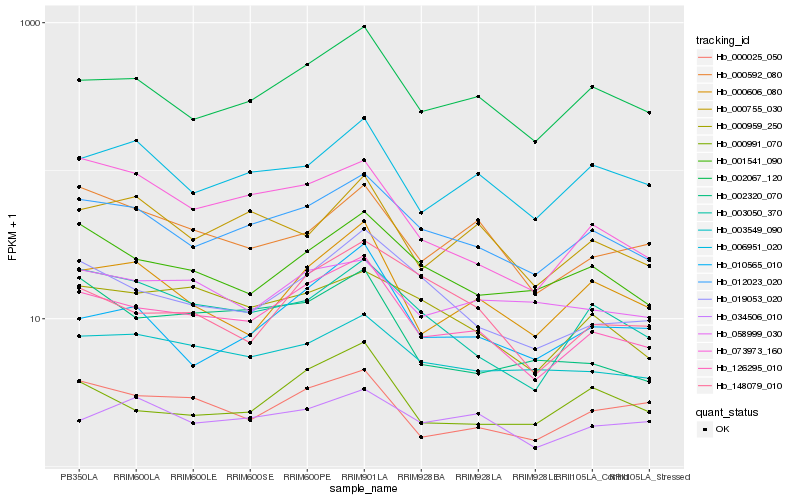
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126295_010 |
0.0 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 2 |
Hb_012023_020 |
0.0853453776 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 3 |
Hb_002067_120 |
0.0866782738 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 4 |
Hb_019053_020 |
0.105003935 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 5 |
Hb_003549_090 |
0.1159936355 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 6 |
Hb_001541_090 |
0.1181502715 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_050 |
0.1181843768 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 8 |
Hb_000606_080 |
0.1190977573 |
- |
- |
PREDICTED: RNA-binding protein Nova-2-like [Malus domestica] |
| 9 |
Hb_000959_250 |
0.119198117 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 10 |
Hb_003050_370 |
0.1205812592 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 11 |
Hb_000991_070 |
0.125336516 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 12 |
Hb_073973_160 |
0.1259482321 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 13 |
Hb_006951_020 |
0.1262778852 |
- |
- |
Rop3 [Hevea brasiliensis] |
| 14 |
Hb_000592_080 |
0.1290786475 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 15 |
Hb_010565_010 |
0.1293111487 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 16 |
Hb_148079_010 |
0.131562426 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 17 |
Hb_058999_030 |
0.1316273154 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_034506_010 |
0.1320740858 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 19 |
Hb_002320_070 |
0.1322106897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000755_030 |
0.1322869261 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |