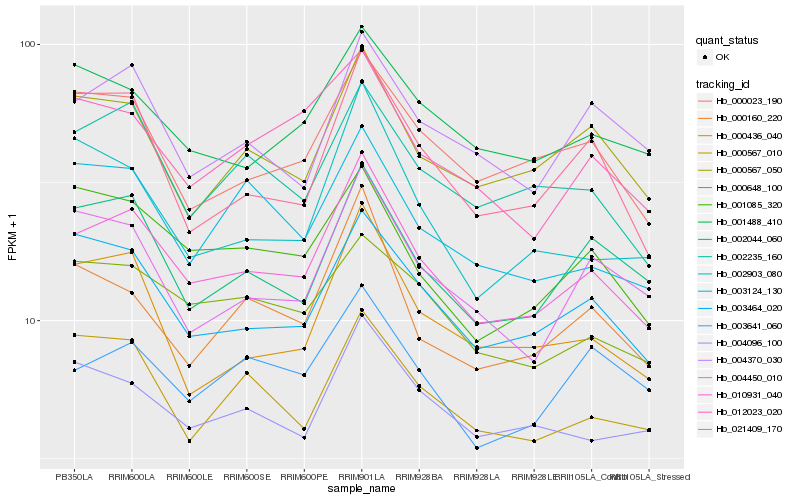
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_040 |
0.0 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 2 |
Hb_003464_020 |
0.0716975618 |
- |
- |
srpk, putative [Ricinus communis] |
| 3 |
Hb_002235_160 |
0.0765616636 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 4 |
Hb_002044_060 |
0.0780565323 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 5 |
Hb_001085_320 |
0.0795331598 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000023_190 |
0.0795493835 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 7 |
Hb_000567_050 |
0.0796544309 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 8 |
Hb_000648_100 |
0.0803743094 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 9 |
Hb_004096_100 |
0.0810755547 |
- |
- |
PREDICTED: NADPH-dependent diflavin oxidoreductase 1 [Jatropha curcas] |
| 10 |
Hb_000160_220 |
0.0822802021 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_001488_410 |
0.0834022696 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 12 |
Hb_012023_020 |
0.0835863721 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 13 |
Hb_004370_030 |
0.085588427 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 14 |
Hb_004450_010 |
0.0858368443 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 15 |
Hb_000436_040 |
0.0863434532 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 16 |
Hb_000567_010 |
0.0865401393 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 17 |
Hb_021409_170 |
0.0865607074 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_003641_060 |
0.0867508225 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 19 |
Hb_003124_130 |
0.08723683 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002903_080 |
0.0893427656 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |