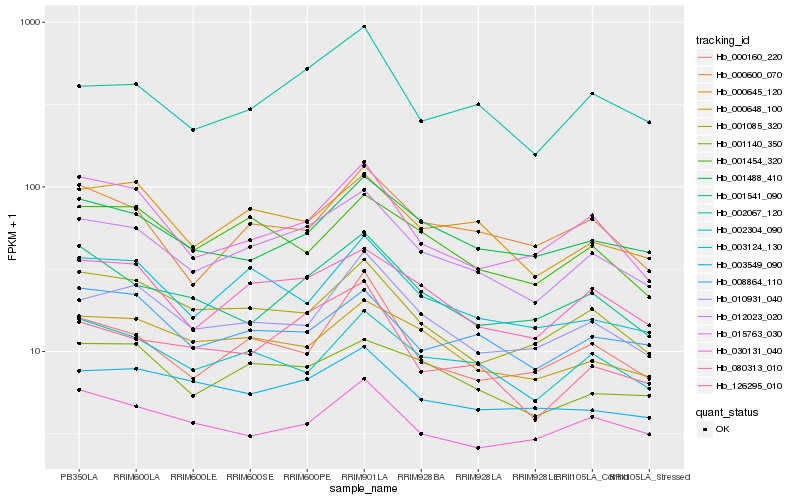
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012023_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 2 |
Hb_080313_010 |
0.0584427086 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 3 |
Hb_002067_120 |
0.076351969 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 4 |
Hb_001140_350 |
0.0828667925 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 5 |
Hb_000645_120 |
0.0831083995 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 6 |
Hb_010931_040 |
0.0835863721 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 7 |
Hb_126295_010 |
0.0853453776 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 8 |
Hb_000648_100 |
0.0861284345 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 9 |
Hb_002304_090 |
0.0865252923 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001488_410 |
0.0869364112 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_001541_090 |
0.0880452778 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003549_090 |
0.0881023097 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 13 |
Hb_001085_320 |
0.0898337001 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_003124_130 |
0.0917512952 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 15 |
Hb_015763_030 |
0.093466097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000600_070 |
0.0936969649 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 17 |
Hb_008864_110 |
0.095582999 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 18 |
Hb_030131_040 |
0.0959666345 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 19 |
Hb_001454_320 |
0.096096551 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000160_220 |
0.0965267271 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |