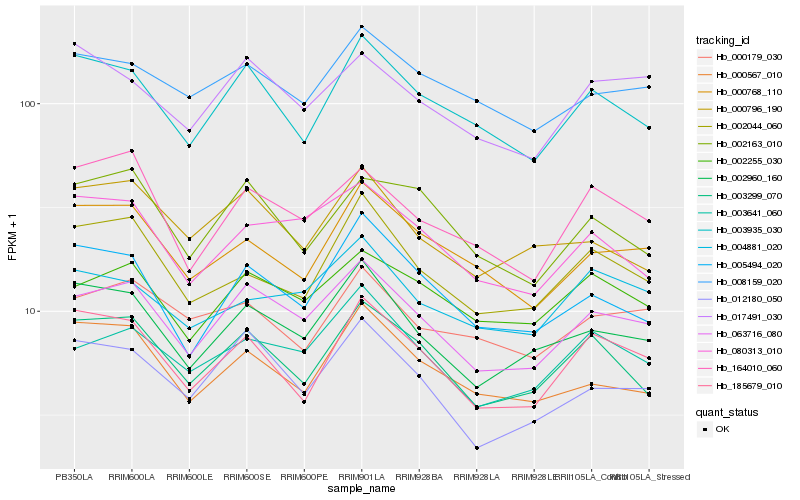
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063716_080 |
0.0 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 2 |
Hb_003641_060 |
0.0661515071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 3 |
Hb_002960_160 |
0.0671896795 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 4 |
Hb_002255_030 |
0.0691484661 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 5 |
Hb_012180_050 |
0.0753930557 |
- |
- |
PREDICTED: uncharacterized protein LOC105632355 isoform X3 [Jatropha curcas] |
| 6 |
Hb_080313_010 |
0.0766608753 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 7 |
Hb_003935_030 |
0.0784601882 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 8 |
Hb_003299_070 |
0.0794737298 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 9 |
Hb_164010_060 |
0.0820948336 |
- |
- |
PREDICTED: uncharacterized protein LOC105633115 [Jatropha curcas] |
| 10 |
Hb_008159_020 |
0.0836921306 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 11 |
Hb_005494_020 |
0.084710861 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 12 |
Hb_000768_110 |
0.0866801737 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_017491_030 |
0.0871718467 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 14 |
Hb_185679_010 |
0.0883908518 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 15 |
Hb_002163_010 |
0.0891658445 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 16 |
Hb_000796_190 |
0.0903258018 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 17 |
Hb_004881_020 |
0.0903474605 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 18 |
Hb_000179_030 |
0.0924968498 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000567_010 |
0.0925552607 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 20 |
Hb_002044_060 |
0.0936569958 |
- |
- |
elongation factor ts, putative [Ricinus communis] |