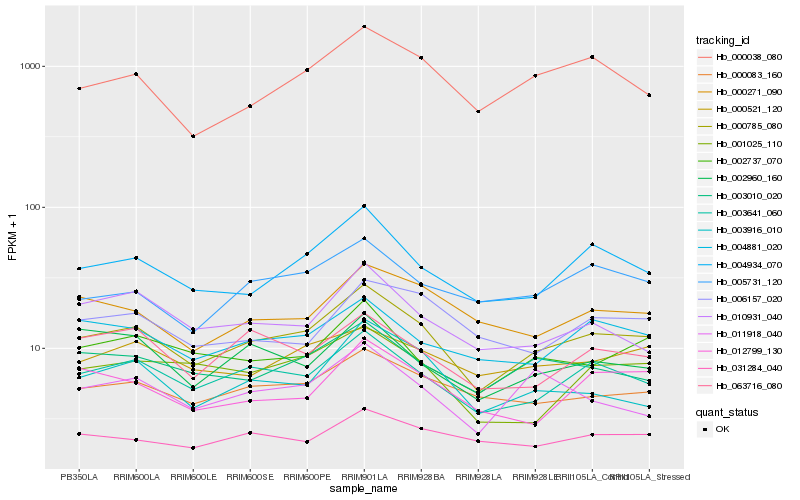
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000785_080 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 2 |
Hb_003641_060 |
0.0722960788 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 3 |
Hb_004934_070 |
0.086314372 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 4 |
Hb_000083_160 |
0.0930798718 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 5 |
Hb_003010_020 |
0.0973889141 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000038_080 |
0.0977760647 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 7 |
Hb_004881_020 |
0.1002044428 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 8 |
Hb_002737_070 |
0.1007022915 |
- |
- |
PREDICTED: phagocyte signaling-impaired protein [Jatropha curcas] |
| 9 |
Hb_005731_120 |
0.1037861838 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP20 [Jatropha curcas] |
| 10 |
Hb_000271_090 |
0.1043773635 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_003916_010 |
0.1044519139 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 12 |
Hb_000521_120 |
0.1077968083 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_063716_080 |
0.1085356309 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 14 |
Hb_031284_040 |
0.1095154021 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 15 |
Hb_012799_130 |
0.1096877106 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 16 |
Hb_001025_110 |
0.1099584281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010931_040 |
0.1105835271 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 18 |
Hb_006157_020 |
0.1131279493 |
- |
- |
PREDICTED: nucleolin 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002960_160 |
0.1136937592 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 20 |
Hb_011918_040 |
0.1139508483 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |