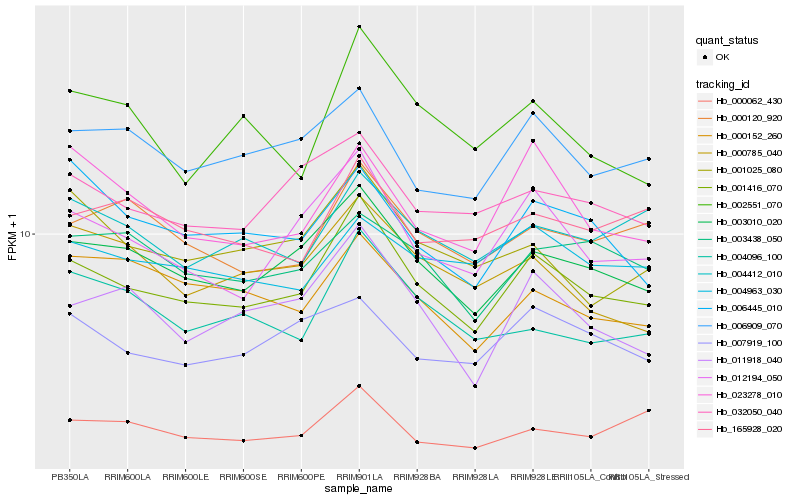
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001416_070 |
0.0 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 2 |
Hb_003010_020 |
0.0587604388 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_012194_050 |
0.0706813135 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 4 |
Hb_011918_040 |
0.0844520978 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_006909_070 |
0.0914965719 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_032050_040 |
0.0936108726 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 7 |
Hb_000120_920 |
0.0954789179 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 8 |
Hb_001025_080 |
0.0963778832 |
- |
- |
hypothetical protein JCGZ_11431 [Jatropha curcas] |
| 9 |
Hb_002551_070 |
0.0965056757 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 10 |
Hb_004096_100 |
0.0987063538 |
- |
- |
PREDICTED: NADPH-dependent diflavin oxidoreductase 1 [Jatropha curcas] |
| 11 |
Hb_007919_100 |
0.1000202734 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 12 |
Hb_004412_010 |
0.100902454 |
- |
- |
- |
| 13 |
Hb_000785_040 |
0.1009780099 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 14 |
Hb_023278_010 |
0.1013638903 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 15 |
Hb_165928_020 |
0.1036287562 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003438_050 |
0.1039858079 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 17 |
Hb_000152_260 |
0.1044203245 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 18 |
Hb_000062_430 |
0.104691843 |
- |
- |
PREDICTED: uncharacterized protein LOC105644764 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006445_010 |
0.1047682311 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 20 |
Hb_004963_030 |
0.104803869 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |