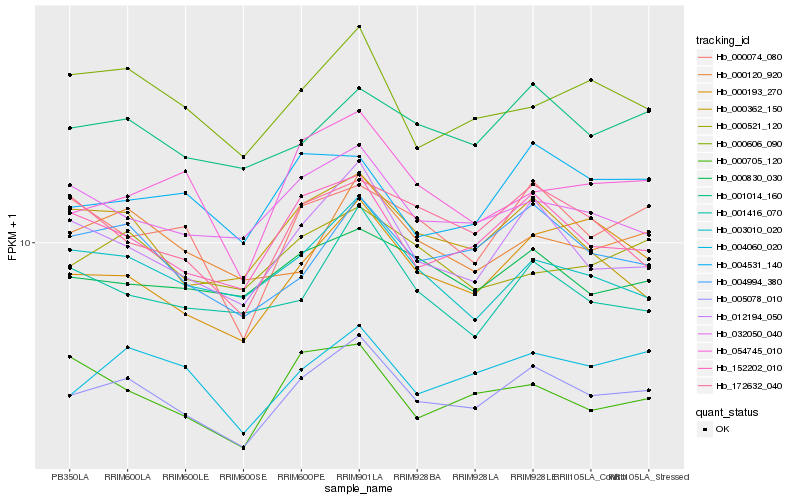
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005078_010 |
0.0 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 2 |
Hb_012194_050 |
0.0798691409 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 3 |
Hb_152202_010 |
0.0889942289 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 4 |
Hb_004060_020 |
0.0968325298 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_004994_380 |
0.0975074722 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 6 |
Hb_000193_270 |
0.0989938858 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000074_080 |
0.1085989032 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_003010_020 |
0.1109824753 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_054745_010 |
0.1119208643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000606_090 |
0.1119672767 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 11 |
Hb_001014_160 |
0.114313774 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 12 |
Hb_000120_920 |
0.1154260564 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 13 |
Hb_000362_150 |
0.1161434211 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 14 |
Hb_032050_040 |
0.116189034 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 15 |
Hb_172632_040 |
0.1183036964 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 16 |
Hb_001416_070 |
0.119543768 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 17 |
Hb_004531_140 |
0.1214060835 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 18 |
Hb_000830_030 |
0.1222684632 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 19 |
Hb_000521_120 |
0.1223093787 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_000705_120 |
0.1230678816 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |