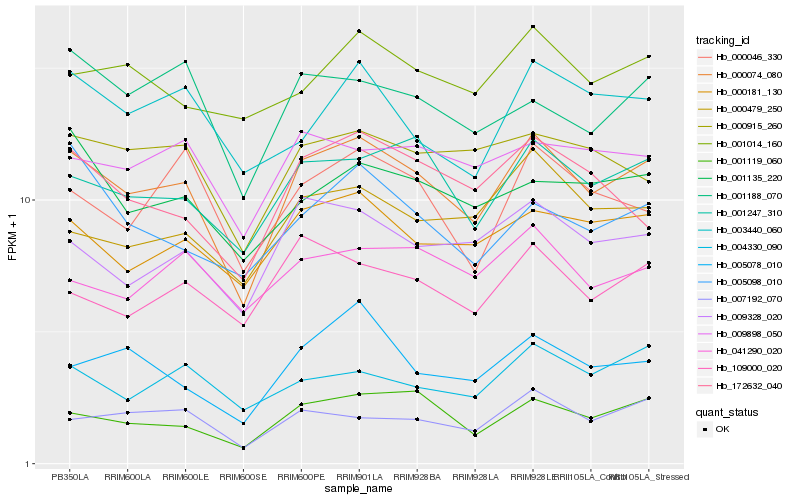
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000074_080 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_001247_310 |
0.0817928107 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |
| 3 |
Hb_001119_060 |
0.0876537219 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 4 |
Hb_000181_130 |
0.089858131 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_009328_020 |
0.0921296758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007192_070 |
0.0957956364 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000915_260 |
0.0995812647 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001188_070 |
0.1000860351 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 9 |
Hb_172632_040 |
0.100686747 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 10 |
Hb_009898_050 |
0.1008411199 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 11 |
Hb_000479_250 |
0.1014122287 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 12 |
Hb_001135_220 |
0.1014549622 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 13 |
Hb_109000_020 |
0.1023679976 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 14 |
Hb_004330_090 |
0.1030365193 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000046_330 |
0.104831285 |
- |
- |
PREDICTED: uncharacterized protein LOC105631826 [Jatropha curcas] |
| 16 |
Hb_003440_060 |
0.1059147476 |
- |
- |
PREDICTED: translin-associated protein X isoform X1 [Jatropha curcas] |
| 17 |
Hb_001014_160 |
0.1060303866 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 18 |
Hb_041290_020 |
0.1070262031 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_005098_010 |
0.1081332416 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [Jatropha curcas] |
| 20 |
Hb_005078_010 |
0.1085989032 |
- |
- |
Derlin-2, putative [Ricinus communis] |