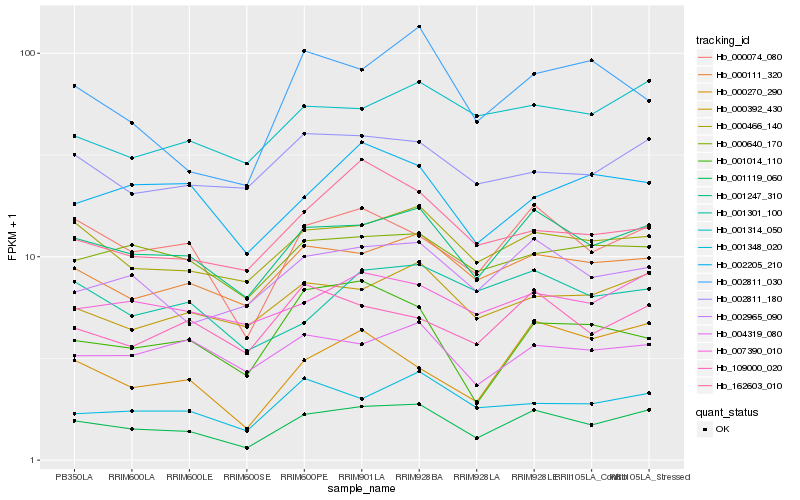
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001119_060 |
0.0 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 2 |
Hb_001247_310 |
0.0669468692 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |
| 3 |
Hb_000074_080 |
0.0876537219 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_000466_140 |
0.1023065655 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 5 |
Hb_000111_320 |
0.1035095242 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 6 |
Hb_002965_090 |
0.108982574 |
- |
- |
importin alpha, putative [Ricinus communis] |
| 7 |
Hb_000392_430 |
0.1105477011 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 8 |
Hb_001348_020 |
0.115383859 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001314_050 |
0.1187469848 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit alpha-like [Jatropha curcas] |
| 10 |
Hb_007390_010 |
0.1199884197 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_002811_180 |
0.121286897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000640_170 |
0.1214766432 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 13 |
Hb_109000_020 |
0.1220878172 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 14 |
Hb_002205_210 |
0.1230882561 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_162603_010 |
0.1239098559 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_002811_030 |
0.124896086 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 17 |
Hb_004319_080 |
0.1252165381 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 18 |
Hb_001301_100 |
0.1252595513 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000270_290 |
0.1260661332 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 20 |
Hb_001014_110 |
0.1262603521 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |