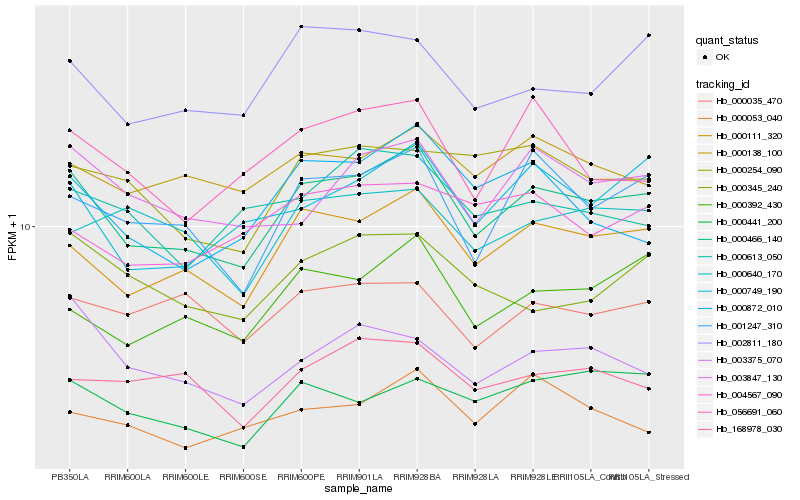
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000466_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 2 |
Hb_000111_320 |
0.045761055 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 3 |
Hb_001247_310 |
0.0656390357 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |
| 4 |
Hb_000392_430 |
0.0684493074 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 5 |
Hb_000749_190 |
0.0694106776 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Populus euphratica] |
| 6 |
Hb_002811_180 |
0.0701652499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003847_130 |
0.0715434165 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 8 |
Hb_000441_200 |
0.0728366554 |
- |
- |
PREDICTED: small RNA 2'-O-methyltransferase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000138_100 |
0.0748929559 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 10 |
Hb_056691_060 |
0.0787613489 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000254_090 |
0.0790568763 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 12 |
Hb_003375_070 |
0.0791136546 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 13 |
Hb_000345_240 |
0.0800646522 |
- |
- |
PREDICTED: cyclin-H1-1 [Jatropha curcas] |
| 14 |
Hb_000053_040 |
0.0802151638 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 15 |
Hb_168978_030 |
0.0804650935 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 16 |
Hb_000872_010 |
0.0814272283 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 17 |
Hb_000035_470 |
0.0818561795 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004567_090 |
0.0823543202 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 19 |
Hb_000613_050 |
0.083743004 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000640_170 |
0.0837664882 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |