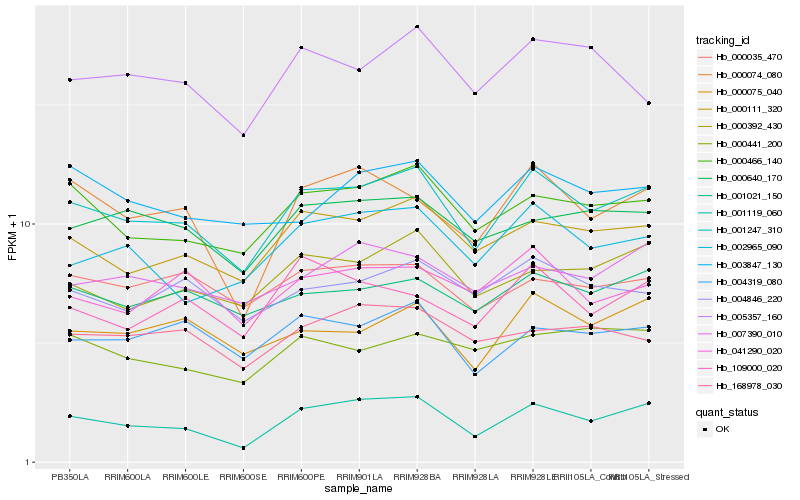
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_310 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |
| 2 |
Hb_000111_320 |
0.0626258064 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 3 |
Hb_000466_140 |
0.0656390357 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 4 |
Hb_001119_060 |
0.0669468692 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X3 [Malus domestica] |
| 5 |
Hb_000075_040 |
0.0760975189 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 6 |
Hb_000392_430 |
0.0776087392 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 7 |
Hb_004319_080 |
0.077997007 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 8 |
Hb_001021_150 |
0.0780500232 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 9 |
Hb_002965_090 |
0.0792343019 |
- |
- |
importin alpha, putative [Ricinus communis] |
| 10 |
Hb_000640_170 |
0.0806323476 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000074_080 |
0.0817928107 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_004846_220 |
0.0819133019 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000035_470 |
0.0840670051 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_041290_020 |
0.084545052 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_003847_130 |
0.0849456094 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 16 |
Hb_007390_010 |
0.0862977386 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_109000_020 |
0.08710955 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 18 |
Hb_005357_160 |
0.0877409788 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 19 |
Hb_000441_200 |
0.089804474 |
- |
- |
PREDICTED: small RNA 2'-O-methyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_168978_030 |
0.0901490252 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |