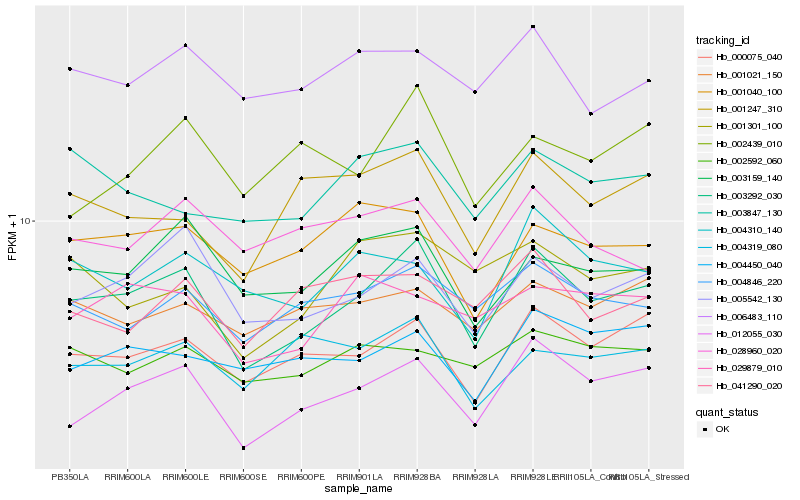
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003292_030 |
0.0 |
- |
- |
PREDICTED: exosome complex component RRP42 [Populus euphratica] |
| 2 |
Hb_012055_030 |
0.0689988097 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 3 |
Hb_000075_040 |
0.0778490119 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 4 |
Hb_005542_130 |
0.0800522718 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 5 |
Hb_003159_140 |
0.0912069671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004846_220 |
0.0917852694 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001247_310 |
0.098382041 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |
| 8 |
Hb_001040_100 |
0.1005414056 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 9 |
Hb_004310_140 |
0.1013712844 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_006483_110 |
0.1018130637 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 11 |
Hb_004450_040 |
0.1018903258 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 12 |
Hb_029879_010 |
0.1026396127 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 13 |
Hb_001301_100 |
0.1041917755 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_001021_150 |
0.1042200756 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 15 |
Hb_003847_130 |
0.1043447427 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 16 |
Hb_004319_080 |
0.1057791218 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 17 |
Hb_041290_020 |
0.1060434608 |
- |
- |
PREDICTED: centromere/kinetochore protein zw10 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_002592_060 |
0.106347538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_028960_020 |
0.1064463912 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002439_010 |
0.1067331022 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |