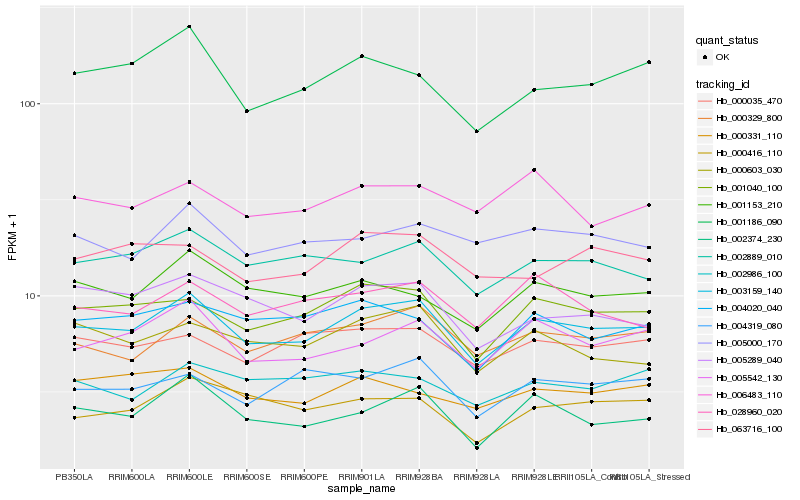
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003159_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001040_100 |
0.0582221099 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 3 |
Hb_000329_800 |
0.065059163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005542_130 |
0.066152104 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 5 |
Hb_002889_010 |
0.0686874918 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 6 |
Hb_001153_210 |
0.0749063459 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000035_470 |
0.0751156093 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000603_030 |
0.0762168543 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 9 |
Hb_001186_090 |
0.0764307009 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 10 |
Hb_002374_230 |
0.0766409747 |
- |
- |
PREDICTED: centromere-associated protein E [Jatropha curcas] |
| 11 |
Hb_028960_020 |
0.0767830422 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005000_170 |
0.0775437932 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 13 |
Hb_005289_040 |
0.0783887428 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 14 |
Hb_004319_080 |
0.0788740942 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 15 |
Hb_004020_040 |
0.0803804821 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 16 |
Hb_063716_100 |
0.0814315336 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000331_110 |
0.081778843 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 18 |
Hb_006483_110 |
0.0820526643 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 19 |
Hb_002986_100 |
0.0830369949 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 20 |
Hb_000416_110 |
0.0831451708 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |